Short Communication Open Access
Detection of Soluble Oligomers Formed by PB1-F2 Influenza A Virus Protein in vitro
Jasmina Vidic *, Ronan Le Goffic, Anna Miodek, Christiane Bourdieu, Charles-Adrien Richard, Mohammed Moudjou, Bernard Delmas, and Christophe Chevalier*
Virologie et Immunologie Moléculaires, Institut National de la Recherche Agronomique, 78350 Jouy en Josas, France
- *Corresponding Author:
- Jasmina Vidic
Virologie et Immunologie Moléculaires
Institut National de la Recherche Agronomique
78350 Jouy en Josas, France
Tel: +0033134652623
Fax: +0033134652621
E-mail: jasmina.vidic@jouy.inra.fr
- Christophe Chevalier
Virologie et Immunologie Moléculaires
Institut National de la Recherche Agronomique
78350 Jouy en Josas, France
Tel: +0033134652623
Fax: +0033134652621
E-mail: christophe.chevalier@jouy.inra.fr
Received date: June 05, 2013; Accepted date: July 20, 2013; Published date: July 23, 2013
Citation: Vidic J, Le Goffic R, Miodek A, Bourdieu C, Richard CA, et al. (2013) Detection of Soluble Oligomers Formed by PB1-F2 Influenza A Virus Protein in vitro. J Anal Bioanal Tech 4:169. doi: 10.4172/2155-9872.1000169
Copyright: © 2013 Vidic J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Influenza A viruses (IAV) remain a major cause of respiratory disease worldwide each year and have been responsible for three main pandemics during the last century comprising the Spanish flu which killed up to 50 million of people. IAV are RNA enveloped viruses belonging to the Orthomyxoviridae family. The nature of the genome of IAV favors the constant and hardly predictable emergence of new strains such as highly virulent H5N1 viruses since 2003, or the H1N1 2009 pandemic strain. Recently, new human cases of severe respiratory illness with a new avian influenza A (H7N9) virus have been reported in China (mortality rate of 60%).Thus, researchers maintain their efforts to determine specific markers of virulence and to evaluate the potential of emerging strains to cause new pandemics.
Influenza A viruses (IAV) remain a major cause of respiratory disease worldwide each year and have been responsible for three main pandemics during the last century comprising the Spanish flu which killed up to 50 million of people. IAV are RNA enveloped viruses belonging to the Orthomyxoviridae family. The nature of the genome of IAV favors the constant and hardly predictable emergence of new strains such as highly virulent H5N1 viruses since 2003, or the H1N1 2009 pandemic strain. Recently, new human cases of severe respiratory illness with a new avian influenza A (H7N9) virus have been reported in China (mortality rate of 60%).Thus, researchers maintain their efforts to determine specific markers of virulence and to evaluate the potential of emerging strains to cause new pandemics.
PB1-F2 is a short polypeptide of approximately 90 amino acids which is dispensable for the viral replication [1]. PB1-F2 is not encoded by all human and swine IAV strains although nearly all the avian IAV express a full length version of the protein suggesting a host-specific function. It is tempting to correlate the mortality rate of a particular viral strain and the expression of PB1-F2 since contemporary H1N1 strains like the pandemic H1N1/2009 virus that no longer express PB1-F2 appeared to be less virulent. Thus, PB1-F2 is considered as one of the viral protein involved in IAV pathogenicity. PB1-F2 displays a number of original features as a short half-life and rapid proteasomedependent degradation, a mitochondrial tropism but also the ability to localize in the nucleus and cytoplasm of infected cells in a cell-typeand viral strain-dependent fashion [2]. The clear function of PB1-F2 is still unknown even if several groups described apoptotic properties that seem to be influenced by multiple factors such as the primary sequence variability and the range of interactions with other viral proteins and host-specific cellular partners [3]. In a previous study, we showed that PB1-F2 could be classified as a member of the intrinsically disordered proteins due to its lack of structure in aqueous solution and its ability to switch from a random to an alpha-helical or a beta-sheet secondary structure [4]. This conformational change could play an important role in the regulation of PB1-F2 interactions with the host proteins during the viral cycle.
There are an increasing number of proteins unrelated to any amyloid pathology that are reported to aggregate in vitro and/or in vivo into polymer assemblies of amyloid type [5]. This feature is probably related to a general tendency of the polypeptide backbone to self-organize into thermodynamically stable polymeric assemblies. Aggregated protein molecules in these fibrillar forms are associated via intra-molecular hydrogen bonds established between the peptide bonds in parallel and anti-parallel beta-strand conformations [6]. In several cases, lipid membranes were shown to favor amyloid fibrillogenesis [5]. We previously shown that negatively charged liposomes in presence of low concentration of recombinant PB1-F2 were destroyed and resulted in amyloid aggregation of PB1-F2 [4]. Furthermore, using Thioflavin S that binds specifically to beta-strand repetitive motives in amyloid fibers, PB1-F2 amyloid structures were detected in membrane vicinities of cells infected with influenza virus [4]. As observed with negatively charged lipids, an anionic detergent, sodium dodecyl sulfate (SDS), in conditions below its critical micellar concentration was shown to induce PB1-F2 aggregation of amyloid fibers [4,7]. In consequence, SDS appears to be a good reagent to trigger and study PB1-F2 oligomerization process in vitro.
The putative role of fibrillated influenza PB1-F2 protein in IAVinfected cells is still unknown and new analytical approaches are needed for its elucidation. In this study, we have tested a new immunosensor as a device for detection of PB1-F2 amyloid oligomers. Rabbit polyclonal anti-oligomer antibody (A11pAb) (In vitrogen) was used as the sensing element since this antibody was previously shown to bind common epitope in amyloids independently to the amino acid side chains [8].
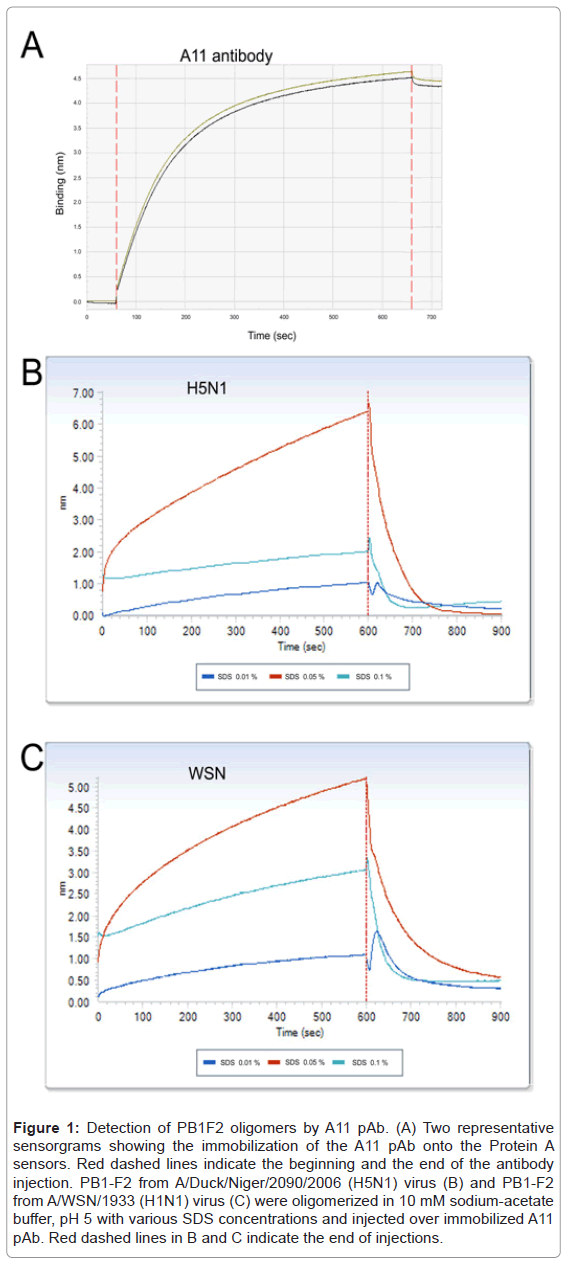
Immunobiosensor experiments were performed on an OctetRED96 platform equipped with protein A sensors (ForteBioInc, Menlo Park, CA). Protein A biosensor (ForteBio, #18-5013) were hydrated in Sample Diluent (ForteBio #18-5028) for 10 min, and then loaded onto the OctetRED96. The A11 pAb was diluted to 25 µg/ml in PBS buffer of pH 7.4 and loaded to saturation after a 60 s baseline in sample buffer. The binding was observed for 600 s. Total of 10 sensors loaded withA11 pAb was prepared for each test. A 4.5 nm shift was observed indicating an efficient immobilization of the antibody (Figure 1A). Finally, the sensors were washed and equilibrated with 10 mM sodium-acetate buffer, pH 5, before probing for PB1-F2 oligomers detection.
Figure 1: Detection of PB1F2 oligomers by A11 pAb. (A) Two representative sensorgrams showing the immobilization of the A11 pAb onto the Protein A sensors. Red dashed lines indicate the beginning and the end of the antibody injection. PB1-F2 from A/Duck/Niger/2090/2006 (H5N1) virus (B) and PB1-F2 from A/WSN/1933 (H1N1) virus (C) were oligomerized in 10 mM sodium-acetate buffer, pH 5 with various SDS concentrations and injected over immobilized A11 pAb. Red dashed lines in B and C indicate the end of injections.
Purified recombinant PB1-F2 proteins from two different influenza virus strains (A/WSN/33 (H1N1) and A/Duck/Niger/2090/2006 (H5N1)) were produced in E. coli and purified as previously described [4]. The two PB1-F2 variants were incubated in SDS solutions of various concentrations ranging from 0 to 0.1% in 10 mM sodium acetate, pH5, buffer for one hour at room temperature to allow oligomerization. The oligomerized PB1-F2 samples were then loaded onto the sensors functionalized with A11 pAb. Figure 1B and 1C show that A11 pAb was able to capture oligomers formed by both PB1-F2 variants in a similar SDS-concentration dependent manner. As expected, no binding was observed in absence of SDS, confirming that A11 pAb does not bind monomeric PB1-F2 (data not shown). The highest sensor signals were recorded in presence of 0.05% SDS for both PB1-F2 variants, while lower signals values were obtained with 0.1 and 0.01% SDS samples. These results are consistent with the Thioflavin T test and the Dynamic Light Scattering measures previously performed indicating that optimal SDS concentration to induce PB1-F2 oligomerization is between 0.01 and 0.05% [4]. A11 pAb was reported to only bind to soluble oligomers independently to the primary sequence and to fail to detect proteins in monomeric form and amyloid fibers [8]. In consequence, our results strongly suggest that PB1-F2 makes soluble amyloid-like oligomers in 0.05% SDS solution. Moreover, PB1-F2 might assemble into soluble oligomers in the vicinity of IAV-infected membranes and tissues. This oligomerized PB1-F2 might contribute to the virulence of IAV since protein soluble amyloid oligomers are inherently cytotoxic [5]. It is admitted that cell membranes are primary targets of amyloid oligomer pathogenesis. This common toxicity mechanism shared by amyloid oligomers can explained previously observed destruction effect of PB1-F2 on artificial membranes that was not evidenced with preformed fibers of PB1-F2 [4].
Besides in silico molecular dynamic stimulations and high resolution imaging techniques, the immunological tools based on conformational antibodies raised against amyloid fibrils and their precursors are of great value to get information on amyloid assemblies. The OctetRED96 platform for detection of PB1-F2 oligomeric forms demonstrated that immunological approach is appropriate for the analysis of protein aggregation to amyloid fibers in a simple and reproducible manner. The major advantage of this assay is the simple, reliable and amenable to high throughput test performance. In addition, there is no need for protein labeling. The assay can be applied to other proteins since A11 pAb detects beta-strand patterns in amyloid oligomers and is not specific to protein primary sequence [8]. A structured protein can shift from a folded to an aggregated state when its stability is reduced. In vitro this can be induced by temperature, pH, and medium polarity, ionic straight or by merely increasing the protein concentration [5]. The aggregation of intrinsically disordered proteins is especially facilitated regarding the flexibility of their structure. An increasing number of papers support the cytotoxicity of aggregates of these kinds of proteins that lack elaborate structure. In consequence, our immunological device is an interesting tool to be applied in cytotoxicological studies. Especially, the immunosensor could be used to allow the detection of PB1-F2 oligomers in lysates of IAV-infected cells and tissues.
Acknowledgements
We thank Dr. Stéphane Biacchesi (INRA, Jouy-en-Josas, France) for critical reading of the manuscript and Mr Turi’s group from Forte bio for their technical help.
References
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, et al. (2001) A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7: 1306-1312.
- Krumbholz A, Philipps A, Oehring H, Schwarzer K, Eitner A, et al. (2011) Current knowledge on PB1-F2 of influenza A viruses. Med Microbiol Immunol 200: 69-75.
- Chakrabarti AK, Pasricha G (2013) An insight into the PB1F2 protein and its multifunctional role in enhancing the pathogenicity of the influenza A viruses. Virology 440: 97-104.
- Chevalier C, Al Bazzal A, Vidic J, Février V, Bourdieu C, et al. (2010) PB1-F2 influenza A virus protein adopts a beta-sheet conformation and forms amyloid fibers in membrane environments. J Biol Chem 285: 13233-13243.
- Stefani M (2012) Structural features and cytotoxicity of amyloid oligomers: implications in Alzheimer's disease and other diseases with amyloid deposits. Prog Neurobiol 99: 226-245.
- Serpell LC, Sunde M, Benson MD, Tennent GA, Pepys MB, et al. (2000) The protofilament substructure of amyloid fibrils. J Mol Biol 300: 1033-1039.
- Vidic J, Chevalier C, Le Goffic R, Bourdieu C, Richard CA, et al. (2013) Surface Plasmon Resonance Immunosensor for Detection of PB1-F2 Influenza A Virus Protein in Infected Biological Samples. J Anal Bioanal Tech S7: 006.
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, et al. (2003) Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300: 486-489.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14011
- [From(publication date):
August-2013 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 9450
- PDF downloads : 4561