Research Article Open Access
Combining Laser-Assisted Microdisstection with/and Immunohistochemistry - RNA Quality of Clinical LCM-Derived Samples
Ewa Malusecka1*#, Anna Fiszer-Kierzkowska1#, Robert Herok1, Stanislaw Wronski2 and Justyna Rembak-Szynkiewicz31Center for Translational Research and Molecular Biology of Cancer, Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology Gliwice Branch, Gliwice, Poland
2Department of Urology, Jan Biziel Memorial University Hospital No 2, Bydgoszcz, Poland
3Radiodiagnostics Department, Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology Gliwice Branch, Gliwice, Poland
#Both the authors have equally contributed
- *Corresponding Author:
- Ewa Malusecka
Center for Translational Research and Molecular Biology of Cancer
Maria Sklodowska-Curie Memorial Cancer Center and
Institute of Oncology Gliwice Branch, Gliwice, Poland
E-mail: maluseck@io.gliwice.pl
Received Date: January 13, 2012; Accepted Date: May 22, 2012; Published Date: May 27, 2012
Citation: Malusecka E, Wska AFK, Herok R, Wronski S, Rembak-Szynkiewicz J (2012) Combining Laser-Assisted Microdisstection with/and Immunohistochemistry - RNA Quality of Clinical LCM-Derived Samples. J Clinic Experiment Pathol 2:112. doi: 10.4172/2161-0681.1000112
Copyright: © 2012 Malusecka E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Clinical & Experimental Pathology
Introduction
Pathological evaluation, based on morphological criteria only, does not explain clinical heterogeneity. Therefore integration of morphological and molecular data is indispensable. However sample material is a mixture of various cell populations - epithelial cells, stromal cells, vasculature-creating cells, and inflammatory cells with variable proportion of various cell types. Consequently analysis of one type cell population is warranted. Microdissection enables obtaining pure cell populations. Moreover microdissection performed on tissue material effects not only in clearing out of unwanted “objects” but also enables to study influence of cell’s surroundings, which is impossible in cultured cells.
Microdissection systems differ by laser type and localization, membrane placement and method of material extraction as well as microdissected material acquiring [1]. Probably due to the price of systems and subsequent analysis their direct comparison has not been done. Microdissection performed on prostate tissue obtained by surgery was frequently described [2-4]. However use of biopsies for microdissection was merely tested [5]. The same is true for other tumor localizations. Limited use of biopsies is connected with small yield of material and the biopsy sampling bias. Nevertheless it has to be remembered that biopsies are sometimes only available study material because they are performed in treatment techniques other than surgery or done before treatment decision. Comparison of data obtained from biopsies and surgical material show that transcription profiles from these samples are similar [6,7].
Samples acquired by microdissection can be analyzed on nucleic acids and/or protein level. To study transcriptome microarray or quantitative PCR are employed. PCR-based methods require less material than microarray and are sensitive enough to be performed on single cell, but the number of genes analyzed is markedly lower.
Our goal was to establish optimal protocol of preparation of slides for microdissection allowing analyzing samples both by immunohistochemistry and RNA analysis. Since RNA quality is influenced by many preserving and fixating compounds we wanted to examine to what extent they influence both IHC and RNA integrity value thus we performed some tests concerning tissue preservation and conditions of microdissection to establish maximum quality of RNA and immunohistochemistry [8].
Materials and Methods
Animal tissue processing
We used mouse tissue (liver, prostate); animals were sacrificed by spine translocation for other purposes. Tissue was frozen using liquid nitrogen, dry ice or RNAlater™ (#AM7024, Ambion). RNA was isolated using RNeasy Mini kit (#74104, Qiagen). In the next step tissue was cryostat cut (Leica CM1950, Leica Microsystems) and slides were fixed in various fixatives.
Human tissue processing
Human samples were prostatectomy specimens and prostate biopsies. Surgical specimens were immediately frozen on dry ice, and stored in deep-freezer in tissue bank. Biopsies were taken from patients with suspected prostate cancer - on the basis of PSA level or DRE examination. Transrectal core biopsies were obtained under USG guidance. From each lobe - four biopsies were used for pathological examination; one additional biopsy was used for molecular analysis. Biopsies for molecular analysis were immediately frozen on dry ice or placed in RNAlater™. Decision on method of biopsy preservation was randomly made. Biopsies for molecular analysis were kept in -700C until use. Pathological diagnosis was known at the time of tissue processing. All samples were obtained after approval of institutional Bioethical Committee.
Before dissection specimens were cryostat cut (Leica CM1950, Leica Microsystems). Cryostat was sterilized with UVC and ethyl alcohol. For every specimen separate knife was used to avoid crosscontamination. Specimens were mounted on the cutting device with OCT compound and immediately frozen with Peltier’s system. Samples were macrodissected from four 10 μm slides. Areas enriched in glandular cells were prepared by removing stromal tissue. For areas enriched in stroma, sections presenting only stromal compartment were used.
Microdissection
Sections subjected to microdissection were mounted on special PEN-coated glass slide (#415101-4401-600, Zeiss), free of nucleases and nucleic acids. Cells were microdissected and further transferred to AdhesiveCaps (#415101-4400-250, Zeiss) by catapulting using PALM system (PALM®MicroBeam, Zeiss), equipped in Olympus microscope. Before microdissection slides were stained with hematoxylin/eosin according to the following protocol: fixation with ice-cold 70% ethyl alcohol - 3 min., water washing, hematoxylin, water washing, Scott’s water - 30 sec., 70% ethyl alcohol - 30 sec., alcohol eosin, 70% ethyl alcohol - 30 sec., 96% ethyl alcohol - 30 sec., 100% ethyl alcohol - 30 sec., 100% ethyl alcohol - 3 min. All dilution were prepared in sterile 50 ml tubes (#62.547.254, Sarstedt) with DEPC- water (#D5758-5ML, Sigma) and handled with gloves. Directly after cutting sections were kept in cryostat chamber, and then transferred to -200C freezer until used for microdissection.
Immunohistochemistry
We tested IHC with AMACR/p63 antibody (#PIN001, PIN cocktail, LOGYBioPrime, Biologo, Germany) on 70% ethyl alcohol-, formalinfixed (1%, 5%, 10%) slides. These sections were mounted on gelatinized glass slides. Briefly - after formalin fixation (20 min in neutral buffered 10% formalin) slides were subjected to antigen retrieval (microwave oven) in citric buffer (pH 6.0; 0.01M). ABC/HRP ABC Vectastain Elite kit (# PK-6200, Vector Labs) was used. Immunohistochemical reaction was performed according to manufacturer instruction, with DAB as a chromogen. Prior to immunohistochemical reaction intrinsic peroxidase was quenched by blocking sections for 10 min in 1% peroxide in PBS. RNase inhibitor (SUPERase In™, Ambion) was added to formalin. It inhibits RNases: A, B, C, 1 and T1.
RNA extraction and analysis
For microarray RNA was isolated using RNeasy Micro kit (#74004, Qiagen), for quantitative RT-PCR RNA was isolated with three different kits RNeasy Micro kit (#74004, Qiagen), Total RNA(# 031-100, A&A Biotechnology), Arcturus PicoPure RNA Isolation Kit (#12204-01, Applied Biosystems). RNA integrity was analyzed using Bioanalyzer 2100 and expressed as RIN number which values range from 10 (intact) to 1 (totally degraded). Four to five cryostat-cut 10 μm sections were taken for RNA isolation.
Quantitative RT-PCR (qRT-PCR)
RNA was converted to ds cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). Two-step cDNA synthesis was applied. Real-Time PCR Master Mix SYBR A (#2008-100A, A&A Biotechnology) was used. Reactions were performed on a CFX real time thermocycler (Bio-Rad)
Microarray analysis
We used U133 Plus 2.0 (Affymetrix) oligonucleotide microarrays. Target preparation from the starting amount of 50 ng RNA, hybridization and staining were done according to Affymetrix instruction manual (Two-Cycle Target Labeling and Control Reagents, Affymetrix and Megascript High Yield Transcription Kit, Ambion were used). Obtained raw microarray data after stringent assessment of hybridization quality (affyQCReport Bioconductor package) were normalized by GC-RMA method [9].
Results
RNA analysis of different tissue preservatives
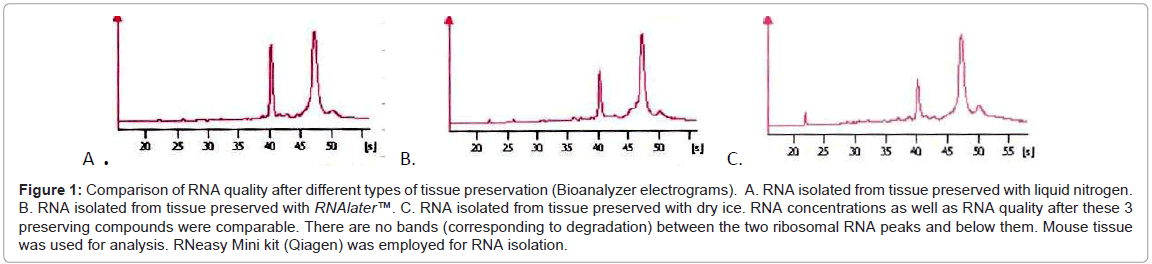
We performed some tests which enables us direct comparison of various methods of tissue preservation, fixation and staining, which is unavailable in routine practice. Tests were done on various mouse tissues, in experimental conditions. First of all we tested the most frequently used tissue preservation agents (liquid nitrogen, dry ice, RNAlater™) on mouse liver and prostate. We obtained RNA of good quality (Figure 1) with all types of tissue preservation agents with quality diminishing slightly in the following order: liquid nitrogen, RNAlater™ and dry ice. Although these differences have to be mentioned, the variations between the three preservatives were small, namely RIN 9.1 (liquid nitrogen) for the best and RIN 8.8 (dry ice) for the worst.
Figure 1: Comparison of RNA quality after different types of tissue preservation (Bioanalyzer electrograms). A. RNA isolated from tissue preserved with liquid nitrogen. B. RNA isolated from tissue preserved with RNAlater™. C. RNA isolated from tissue preserved with dry ice. RNA concentrations as well as RNA quality after these 3 preserving compounds were comparable. There are no bands (corresponding to degradation) between the two ribosomal RNA peaks and below them. Mouse tissue was used for analysis. RNeasy Mini kit (Qiagen) was employed for RNA isolation.
Influence of hematoxylin staining on RNA quality
To test whether and to what extent hematoxylin staining influences RNA quality we compared unstained and hematoxylin-stained tissue. Mouse prostate and liver in RNAlater™ and dry ice was cryostat-cut, fixed, stained but not microdissected. Results of this experiment show that hematoxylin staining degrades RNA quality only slightly – RIN=7 of unstained sample, RIN=6.7 of hematoxylin stained samples (not shown).
RNA analysis of dissected human prostate
To test whether microdissection itself influences RNA quality we performed comparison of the three sections from the same prostatectomy specimens: 1. cut only, 2. after fixation with 70% ethyl alcohol and 3. after microdissection. We found out that the process of microdissection diminishes RNA integrity roughly by the factor of 1 (microdissected cells as compared to cut only), absolute values depending of the RIN of starting material (not shown, data available on request).
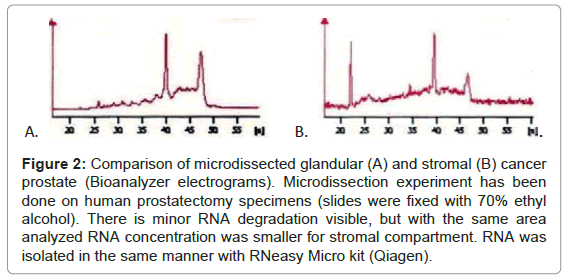
In the next step we compared the amount of RNA obtained from glandular and stromal prostate cells. At first, we macrodissected two areas - epithelial (rich for prostate glands) and stromal (devoid of epithelial cells). Macrodissection was done on prostatectomy specimens. With comparable areas (~0.5 cm2) and RIN values (~7) we obtained more than double RNA from epithelial fragment than from stromal one. Secondly, we microdissected stromal and glandular cells. Microdissection was done on cancerous and non-cancerous parts of prostatectomy specimens. With almost threefold larger cut area (1.3 million μm2 for glands vs. 3.5 million μm2 for stroma) we obtained less RNA and the quality of RNA was slightly worse for stromal compartment (Figure 2, exemplary RIN and concentration –control (whole) tissue RIN= 7.2, conc.=21ng/μl; microdissected glandular cells RIN=6.8, conc.= 3.8 ng/μl; microdissected stromal cells RIN= 6.0, conc.=2.2 ng/μl). These results demonstrate that amounts of RNA obtained from stromal cells (macro- or microdissected) are always reduced in comparison with glandular cells./p>
Figure 2: Comparison of microdissected glandular (A) and stromal (B) cancer prostate (Bioanalyzer electrograms). Microdissection experiment has been done on human prostatectomy specimens (slides were fixed with 70% ethyl alcohol). There is minor RNA degradation visible, but with the same area analyzed RNA concentration was smaller for stromal compartment. RNA was isolated in the same manner with RNeasy Micro kit (Qiagen).
RNA quality in formalin fixed tissue
We performed some test on RNA quality and IHC, as our aim was to combine microdissection and RNA analysis. These tests were performed on non-microdissected human prostate biopsies to avoid interference with reduced RNA content. We employed AMACR/p63 staining to identify glandular and basal cells.
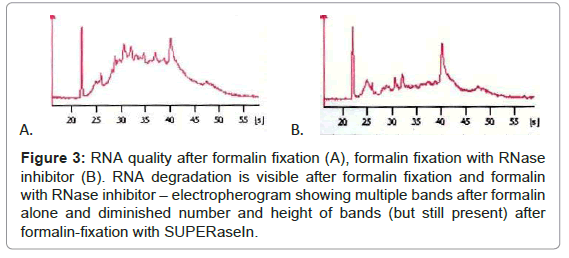
While immunohistochemical reaction after formalin-fixation was good, RNA quality was insufficient, thus we tested if diminishing percentage of formalin influence the staining effectiveness or RNA quality. Immunohistochemistry with 1% formalin fixation and diminished antibodies incubation time was excellent, but RNA quality was poor, the same as with higher formalin percentages (Figure 3). Addition of RNase inhibitor (SUPERase In™, Ambion) makes RNA quality slightly better, but RNA degradation was still clear. RNA bands of different sizes corresponding to RNA degradation were present after formalin fixation (RIN=3.3), irrespectively of adding the inhibitor (RIN=5.0) (Figure 3).
Figure 3: RNA quality after formalin fixation (A), formalin fixation with RNase inhibitor (B). RNA degradation is visible after formalin fixation and formalin with RNase inhibitor – electropherogram showing multiple bands after formalin alone and diminished number and height of bands (but still present) after formalin-fixation with SUPERaseIn.
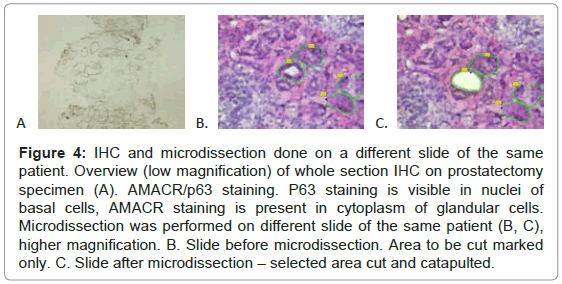
Taking into account these results we decided to perform IHC staining and RNA analysis on separate slides. IHC sections were mounted on gelatinized glass slides. We decided to stain with AMACR/ p63 antibody and formalin-fix first, middle and last slide and perform microdissection and RNA analysis on neighboring, 70% ethyl alcohol fixed, hematoxylin stained dry-ice or RNAlater™ preserved tissue slides (Figure 4).
Figure 4: IHC and microdissection done on a different slide of the same patient. Overview (low magnification) of whole section IHC on prostatectomy specimen (A). AMACR/p63 staining. P63 staining is visible in nuclei of basal cells, AMACR staining is present in cytoplasm of glandular cells. Microdissection was performed on different slide of the same patient (B, C), higher magnification. B. Slide before microdissection. Area to be cut marked only. C. Slide after microdissection – selected area cut and catapulted.
Quantitative RTPCR and microarray analysis of LCM samples
In order to analyze gene expression in glandular and stromal prostate cells [10-14] we performed microdissection of selected cells followed by microarray and/or quantitative RT-PCR. Since this system shows only the size of the cut area, we were not able to establish precisely number of analyzed cells. In case of glandular cancer cells acquired area ranged from 2410 – 298088 μm2. Microdissection was done in 30 cases. We tested three different kits (Qiagen RNeasy Micro kit, A&A Biotechnology kit, and Arcturus PicoPure RNA Isolation Kit) for isolation of RNA from microdissected material. Two of them (Qiagen RNeasy Micro kit, and A&A Biotechnology kit) were not successful for small samples. With Arcturus PicoPure RNA Isolation Kit we obtained enough RNA to perform PCR reaction on one gene. Concentration of isolated material was 2-5 ng/μl. The received results of RNA concentration did not correlate with the size of microdissected area. We are aware that within this range measurement of RNA concentration is unreliable.
As the results of QRT-PCR analysis of microdissected cancer cells were unsatisfactory we performed two separate experiments. We microdissected substantially larger areas (`~2 millions μm2) – stromal cells were cut from prostatectomy material. These/stromal cells were chosen to achieve larger areas of microdissection which were unavailable with cancer glandular cells not for its transcriptional profiling. We obtained on average 25 ng of total RNA with RIN value ranging between 5 and 6. RNA content and quality was of enough good to perform DNA microarrays. Fifteen samples obtained this way were subjected for two rounds of amplification yielding on average 19 μg labeled cRNA. Hybridization of 14 samples with microarrays was successful. Hybridization results were stringently assessed by affyQC Report Bioconductor package and the quality of obtained raw microarray data was valued as good.
On the other hand, we examined RNA quality of nonmicrodissected, small tissue fragment. Analysis of RNA integrity and concentration of tissue small fragment (~1 mm3) showed that RIN value as well as concentration was markedly reduced as compared to parental, bigger fragment (RIN 1.3 vs.5.2; concentration 6.7 ng/μl vs. 39 ng/μl). All other factors connected with isolation were not changed. Examination of small fragment of tissues and microdissected areas led us to conclusion that material abundance is major aspect influencing results.
Discussion
Studies combining IHC and nuleic acids analysis with the use of human samples require resolving some technical issues concerning methods preservation, fixation and staining of specimens. As far as preserving of tissue is concerned, all types of preservatives have pros and cons. Freezing in liquid nitrogen is rapid, and gives excellent results for RNA analysis and IHC. But it has to be remembered that liquid nitrogen evaporates quickly so it is challenging in everyday use, in nonexperimental conditions. Dry ice is very good tissue preservative, sample freezing is relatively fast as compared to liquid nitrogen as the standard and very good IHC staining. Both of aforementioned chemicals do not fix tissue but only preserve it therefore sample must be frozen all the time or it degrades. On the other hand RNAlater™ can be aliquoted and kept until use. Storage of RNAlater™ samples is easy till they can be kept in the domestic refrigerator. Handling of RNAlater™ tissue is simple since it penetrates the specimen and protects RNA from degradation, even in room temperature for one week (producer statement). However cryostat sectioning of RNAlater™ specimens is difficult and morphology of sections is poor, moreover immunohistochemistry of these slides is more complex. Washing of RNAlater™ improves the cutting and IHC staining but abolishes preservative effect of RNAlater™ [15,16]. Therefore in our opinion dry ice is the best solution for routine use. Use of formalin-fixed tissue for RNA analysis is completely different issue.
There are differences between IHC and RNA analysis in terms of optimal tissue processing. Mostly these dissimilarities are connected with water presence (water diminishes RNA quality) and sterility (IHC does not requires sterilization of fluids and consumables).We show that formalin fixation, although valuable for IHC, is deleterious for RNA quality (which was not surprising) and diminishing formalin concentration and time of fixation does not change much. Masuda et al. [11] have shown that RNA after formalin fixation is not only degraded but also chemically modified. As a consequence results of RNA analysis of formalin-fixed issue should not be compared with unfixed tissue, what is frequently practiced [18,19]. Comparison of formalin-fixed and fresh-frozen breast samples show marked difference in those two types of samples [20] Summarizing our and other experience [21]. We consider formalin-fixed tissue as not the optimal source of RNA for next analysis. We think that our strategy – slides designed for histological of IHC analysis fixed with formalin, and slides fixed with alcohol-based fixative for RNA analysis- is better.
We used antibody which requires formalin fixation and antigen retrieval for successful staining however use of diverse antibodies does not exclude different fixation method, less harmful for RNA.
Another issue which should be taken into account is stromal involvement. We and others [3,22]. show, that RNA yield vary greatly between tissue compartments. Both in macro- and microdissection of glandular cells amount of RNA is much greater than in case of stromal areas. This phenomenon might be connected with diminished nucleus/cytoplasm ratio and/or different transcriptomal activity of stromal compartment. Results presented by de Bruin [22] demonstrate that presence of stromal cells had minor effect on RNA profiling, in contrary to additional rounds of amplification. Apart from completely different expression profiles, admixture of limited number of stromal cells has no major impact on global expression.
Review of papers dealing with analysis of microdissected samples shows that application of this method solve some problems (e.g. cells heterogeneity) but new issues arise (e.g. material scarcity).
References
- Ladanyi A, Sipos F, Szoke D, Galamb O, Molnar B, et al. (2006) Laser microdissection in translational and clinical research. Cytometry A 69: 947-960.
- Hellwinkel OJ, Asong LE, Rogmann JP, Sültmann H, Wagner C, et al. (2011) Transcription alterations of members of the ubiquitin-proteasome network in prostate carcinoma. Prostate Cancer Prostatic Dis 14: 38-45.
- Gregg JL, Brown KE, Mintz EM, Piontkivska H, Fraizer GC (2010) Analysis of gene expression in prostate cancer epithelial and interstitial stromal cells using laser capture microdissection. BMC Cancer 10: 165.
- Tomlins SA, R Mehra, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, Shah RB, and Chinnaiyan AM. (2006) Integrative molecular concept modeling of prostate cancer progression Nature Genetics. 39: 41-51
- Qian DZ, Huang CY, O'Brien CA, Coleman IM, Garzotto M, et al. (2009) Prostate cancer-associated gene expression alterations determined from needle biopsies. Clin Cancer Res 15: 3135-3142.
- Sotiriou C, Chand K, Petersen D, Jazaeri AA, Liu ET (2001) Core biopsy versus surgical tumor specimens for microarray analysis of gene expression profiles Nature Genetics. 27: 88-89.
- Zanetti-Dällenbach R, Vuaroqueaux V, Wight E, Labuhn M, Singer G, et al. (2006) Comparison of gene expression profiles in core biopsies and corresponding surgical breast cancer samples. Breast Cancer Res 8: R51.
- Sample Pooling for Microarray Analysis: A Statistical Assessment of Risks and Biases. Affymetrix Technical Note
- Leiva IM, Emmert-Buck MR, Gillespie JW (2003) Handling of clinical tissue specimens for molecular profiling studies. Curr Issues Mol Biol 5: 27-35.
- Hewitt SM, Lewis FA, Cao Y, Conrad RC, Cronin M, et al. (2008) Tissue handling and specimen preparation in surgical pathology: issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissue. Arch Pathol Lab Med 132: 1929-1935.
- Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K (1999) Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res 27: 4436-4443.
- Farragher SM, Tanney A, Kennedy RD, Paul Harkin D (2008) RNA expression analysis from formalin fixed paraffin embedded tissues. Histochem Cell Biol 130: 435-445.
- Abdueva D, Wing M, Schaub B, Triche T, Davicioni E (2010) Quantitative expression profiling in formalin-fixed paraffin-embedded samples by affymetrix microarrays. J Mol Diagn 12: 409-417.
- Oberli A, Popovici V, Delorenzi M, Baltzer A, Antonov J, et al. (2008) Expression profiling with RNA from formalin-fixed, paraffin-embedded material. BMC Med Genomics 1: 9.
- Srinivasan M, Sedmak D, Jewell S (2002) Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol 161: 1961-1971.
- de Bruin EC, van de Pas S, Lips EH, van Eijk R, van der Zee MM, et al. (2005) Macrodissection versus microdissection of rectal carcinoma: minor influence of stroma cells to tumor cell gene expression profiles. BMC Genomics 6: 142.
- Goldsworthy SM, Stockton PS, Trempus CS, Foley JF, Maronpot RR (1999) Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog 25: 86-91.
- King C, Guo N, Frampton GM, Gerry NP, Lenburg ME, et al. (2005) Reliability and reproducibility of gene expression measurements using amplified RNA from laser-microdissected primary breast tissue with oligonucleotide arrays. J Mol Diagn 7: 57-64.
- Klee EW, Erdogan S, Tillmans L, Kosari F, Sun Z, Wigle DA, Yang P, Aubry MC, and Vasmatzis G. ( 2009) Impact of sample acquisition and linear amplification on gene expression profiling of lung adenocarcinoma: laser capture micro-dissection cell-sampling versus bulk tissue-sampling. BMC Med Genomics. 2: 13.
- Kube DM, Savci-Heijink CD, Lamblin AF, Kosari F, Vasmatzis G, et al. (2007) Optimization of laser capture microdissection and RNA amplification for gene expression profiling of prostate cancer. BMC Mol Biol 8: 25.
- Li Y, Ali S, Philip PA, Sarkar FH (2003) Direct comparison of microarray gene expression profiles between non-amplification and a modified cDNA amplification procedure applicable for needle biopsy tissues. Cancer Detect Prev 27: 405-411.
- Taylor TB, Nambiar PR, Raja R, Cheung E, Rosenberg DW, et al. (2004) Microgenomics: Identification of new expression profiles via small and single-cell sample analyses. Cytometry A 59: 254-261.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15038
- [From(publication date):
June-2012 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 10336
- PDF downloads : 4702