Research Article Open Access
Chronic Prenatal Ethanol Exposure Disrupts WNT Signaling In Adolescent Cerebella
Fusun Gundogan4,6, Ming Tong2,3,6, Mai He4,6, William Cy Chen2,3, Charles Kim5, Quynh-Giao Nguyen2,3, Rosa Yu5 and Suzanne M. de la Monte1-3,6*1Departments of Pathology (Neuropathology), Neurology, Neurosurgery, and Medicine, USA
4Department of Pathology of the Women and Infants Hospital of Rhode Island, USA
6Warren Alpert Medical School, Providence, RI, USA
- *Corresponding Author:
- Suzanne M. de la Monte
Rhode Island Hospital
55 Claverick Street, 4th Floor
Providence, RI 02903, USA
Tel: 401-444-7364
Fax: 401-444-2939
E-mail: Fusun_Gundogan@Brown.edu; Suzanne_DeLaMonte_MD@Brown.edu
Received Date: April 12, 2013; Accepted Date: August 14, 2013; Published Date: August 16, 2013
Citation: Gundogan F, Tong M, He M, Chen WC, Kim C, et al. (2013) Chronic Prenatal Ethanol Exposure Disrupts WNT Signaling In Adolescent Cerebella. J Clin Exp Pathol 3:144. doi: 10.4172/2161-0681.1000144
Copyright: © 2013 Gundogan F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Clinical & Experimental Pathology
Abstract
Background: In Fetal Alcohol Spectrum Disorder (FASD), structural and functional abnormalities in the cerebellum persist through adolescence and beyond. We hypothesize that perturbations in Wnt signaling may contribute to these effects because Wnt pathways mediate neuronal morphogenesis, migration, and plasticity during development.
Objectives: We utilized an established model of FASD to assess the nature and degree to which chronic prenatal ethanol exposure impairs Wnt pathway gene expression in postnatal and adolescent cerebella. Methods: Pregnant Long Evans rats were fed isocaloric liquid diets containing 0%, 18% or 37% ethanol by caloric content from gestation day 6 through delivery. Offspring were sacrificed on postnatal day 3 (P3), P10, P20, or P35. Wnt pathway gene expression was examined using a targeted PCR array and duplex qRT-PCR analysis. Results: Among the 84 genes examined by targeted array, chronic prenatal ethanol exposure (37%) down-regulated just 3 genes at P10, but 33 genes at P35. Further analysis focusing on Wnt5a, Wnt5b, Fzd4, Fzd6, Axin2, Dixdc, and EP300 revealed that prenatal ethanol exposure enhanced expression of Fzd4, Fzd6, Axin2, Dixdc, and EP300 at P3 and Wnt5b at P20, but inhibited Fzd4 and EP300 at P10, and Fzd4, Fzd6, Wnt5a, and EP300 at P20. In addition, ß-catenin, Cyclin D1, and c-Myc protein expression was inhibited by ethanol exposure at P3, but not P10 or P20. To some extent responses were ethanol dose-dependent.
Conclusions: The consequences of chronic prenatal ethanol exposure on Wnt signaling in
the brain shift dramatically with developmental stage. The findings further suggest that Wnt functions such as plasticity, which are needed for adolescent brain development, should be therapeutically targeted in FASD.
Keywords
Fetal alcohol spectrum disorder; Wnt; Cerebellum; Prenatal ethanol exposure
Introduction
Alcohol misuse during pregnancy causes fetal alcohol spectrum disorder (FASD) [1,2], which includes neurodevelopmental abnormalities such as cerebellar hypoplasia and motor function deficits. Ethanol’s inhibitory effects on the immature Central Nervous System (CNS) are mediated in part by inhibition of insulin and insulinlike growth factor (IGF) signaling [3]. Insulin and IGF regulate a broad array of cellular functions in the brain, including neuronal survival and differentiation, myelin formation and maintenance, neuronal migration, plasticity, metabolism, and neurotransmitter supply and responsiveness [4-12]. Therefore, inhibitory effects of ethanol on insulin/IGF signaling could explain many of the neurodevelopmental abnormalities in FASD [1,13-18]. On the other hand, growing evidence suggests that the consequences of impaired insulin/IGF signaling may be indirect and mediated by cross-talk with other pathways, including Wnt and Notch [19,20]. Wnt is of particular interest because to its demonstrated roles in development and neuronal plasticity.
Wnts comprise a family of cysteine-rich secreted signaling glycoproteins that promote diverse physiological functions through coordinated interactions with Frizzled receptors and transcription factors. Wnt signaling is transmitted through three major pathways: canonical, Wnt/Calcium, and planar cell polarity [21-23], and activated when Wnt ligands bind to Frizzled receptors (Fzd), which are 7-transmembrane-spanning G protein-coupled proteins. Secreted or soluble Fzd-related proteins can also interact with Wnts to modulate function, including by inhibition. During development, Wnts mediate cell fate, intercellular adhesion, migration, and proliferation [24,25]. In maturing and adult brains, Wnts regulate neurogenesis and synapse formation which are needed for plasticity, cognitive function, and activity-dependent learning.
In the absence of Wnt ligands, Wnt signaling is constitutively inhibited by GSK-3β-mediated phosphorylation of β-catenin, which targets β-catenin for degradation via the ubiquitin-proteasome pathway. GSK-3β phosphorylation of β-catenin is facilitated by the formation of a cytosolic destruction complex composed of Axin, adenomatous polyposis coli (APC), GSK-3β, and β-catenin. Wnt is activated upon binding of Wnt ligands to Fzd receptors and attendant interaction with LRP5/6 co-receptors which help recruit and activate disheveled. Activated disheveled inhibits GSK-3β, preventing Ser and Thr phosphorylation and subsequent degradation of β-catenin. Stabilized β-catein accumulates in the cytosol and translocates to the nucleus where it interacts with the transcription factors, T Cell Specific Factor (TCF) and Lymphoid enhancer binding factor 1 (LEF-1), to regulate (dis-inhibit) expression of target genes such as Cyclin D, Myc, Axin-2, MMP-7, CD4, and PPAR-δ.
Canonical Wnt signaling is positively supported by the LRP5/6 co-receptors, Dishevelled (Dsh), Akt, GBP/Frat1, CK1, and Rac1, and negatively regulated by secreted/soluble Frizzled proteins, Wnt Inhibitory factor 1 (WIF-1), Dkk, Kremen, and Naked. The Wnt/ Calcium pathway is regulated by G-coupled phospholipases and proteins. Stimulation of this pathway increases cytoplasmic free calcium, and activates signaling through protein kinase C, calcium calmodulin-mediated kinase II (CK2), and calcineurin (phosphatase) [22,26]. The Wnt planar cell polarity pathway is activated by the RAS homologue gene family member A (RhoA) and RAC1, leading to stimulation of stress kinases such as Jun N-terminal kinase (JNK) and RHO-activated coil-containing protein kinase 1 (ROCK) [27-31]. Attendant remodeling of the cytoskeleton alters cell adhesion and motility.
The potential role of impaired Wnt signaling as a mediator of CNS developmental and long-term abnormalities in FASD was drawn into focus by evidence that: 1) ethanol impairs insulin/IGF signaling in the brain; 2) insulin/IGF networks cross-talk with Wnt [19,32-35]; 3) both canonical Wnt and insulin/IGF networks regulate GSK-3β [36,37], and 4) alcohol disrupts Wnt signaling and gene expression [38,39], including in immature neuronal cells [40-42]. Moreover, like insulin/ IGF networks, Wnts regulate CNS neuronal proliferation, migration, differentiation, and axon outgrowth [43-46]. Previous studies showed that prenatal ethanol exposure causes sustained deficits in insulin/IGF signaling with increased activation of GSK-3β in postnatal cerebella [47,48]. Increased levels of GSK-3β inhibit Wnt by increasing β-catenin phosphorylation, ubiquitination, and proteasome degradation [20,23,49-51]. However, little is known about the effects of prenatal ethanol exposure on Wnt signaling in the developing/maturing brain. Herein, we utilized an established experimental model of FASD to characterize effects of chronic moderate- and high-dose prenatal ethanol exposures on Wnt gene expression in cerebella of postnatal (P) day 3, P10, P20, and P35 rats.
Materials and Methods
Materials
Qiazol reagent, EZ1 RNA universal tissue kit, QuantiTect SYBR Green polymerase chain reaction (PCR) master mix, the BIO Robot Z1, and Wnt RT2 Profiler PCR Array System were from Qiagen Inc (Valencia, CA). The AMV first strand cDNA synthesis kit, ProbeFinder Version 2.45 software for design of primer-probe pairs, and LightCycler 480 Real-Time PCR System and software were from Roche Diagnostics Corporation (Indianapolis, IN). Diets used to generate FASD models were purchased from BioServ (Frenchtown, NJ).
Ethanol exposure model
Pregnant Long Evans rats were pair-fed with isocaloric liquid diets in which ethanol comprised 0%, 18%, or 37% of the caloric content. Pregnant rats were fed with liquid diets from gestation day (GD) 6 until parturition [52,53]. GD 0 was when sperm was first detected in vaginal smears. GD6 was selected as the starting point for ethanol feeding because earlier exposures lead to excessive fetal loss [54]. Pregnant dams were monitored daily to ensure equivalent caloric intake and body weight maintenance. After birth, maternal diets were changed to standard chow. Pups were monitored daily and weighed weekly. At sacrifice, freshly harvested cerebella were immediately snap frozen in a dry ice-methanol bath and stored at –80°C. The cerebellum was studied because it is a major target of ethanol induced neurotoxicity [1-3]. The Lifespan-Rhode Island Hospital IACUC committee approved these procedures and the use of rats in experiments.
PCR Array studies
The RT2 Profiler PCR Array System was used to examine the effects of chronic prenatal ethanol exposure on Wnt pathway signaling in the cerebellum. To generate cDNA templates, frozen cerebellar tissues were homogenized in Qiazol reagent, and RNA isolated using the RNeasy Mini kit was reverse transcribed using the AMV First Strand cDNA synthesis kit and random oligodeoxynucleotide primers. Cerebella from 4 groups were studied: P10 and P35 controls, and P10 and P35 ethanolexposed (37% diet) offspring. For each group, 9 different cerebella were used to prepare 3 unique balanced cDNA template pools (from 5 µg of total RNA). The cDNAs were added to SYBR Green based RT2 qPCR Master Mix and dispensed across commercially prepared PCR arrays in 96-well plates. Each well contained a single primer pair, and each plate included the 5 housekeeping standardized control reactions for normalizing results. Reactions were performed in technical triplicates. The RT2 Profiler PCR Array System Excel-based template was used to calculate fold differences (ΔΔCt method) between control and ethanolexposed cerebella.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) analysis
We used qRT-PCR analysis to measure mRNA expression of selected genes that were informative in the RT2 Profiler PCR Arrays. Primer-probe pairs were designed using ProbeFinder Version 2.45 software, and target specificity was verified using NCBI-BLAST (Basic Local Alignment Search Tool). Amplified signals were detected and analyzed using the LightCycler 480 Real-Time PCR System and software. Gene expression was normalized to β-actin which was measured simultaneously in duplex qPCR reactions. Inter-group comparisons were made using calculated mRNA/β-actin ratios.
Duplex enzyme-linked immunosorbent assay (ELISA)
We used duplex direct-binding ELISAs to measure immunoreactivity to b-catenin, Cyclin D1, and C-Myc, which are targets of Wnt signaling. Immunoreactivity was normalized to large ribosomal protein (RPLPO) [15]. Fluorescence was measured in a SpectraMax M5 (Molecular Devices, Sunnyvale, CA). Binding specificity was determined from negative control incubations with primary or secondary antibody omitted. Calculated ratios of specific protein/RPLPO immunoreactivity were used for inter-group comparisons.
Statistical analysis
Boxplots depict means (horizontal bars), 95% confidence intervals (box limits), and range (whiskers). Inter-group comparisons were made using two-way analysis of variance and post hoc Tukey tests with GraphPad Prism 6 software (San Diego, CA).
Results
Targeted array results
Targeted PCR arrays were used to discover the breadth of Wnt pathway abnormalities in cerebella of juvenile (P10) and young adolescent (P35) rats that had been subjected to chronic prenatal ethanol exposure. 84 target and 5 housekeeping genes were measured. The genes, their related Wnt pathways (canonical, planar cell polarity or Wnt/Ca2+), and cellular functions are listed in Supplementary Table 1. Gene expression was measured in 3 sets of pooled samples per group. The results were normalized to Hprt1 because its expression was unchanged with respect to age and ethanol exposure.
| Symbol | 2^-ΔCtEthanol | 2^-ΔCtControl | Fold Change | P-value |
|---|---|---|---|---|
| Apc | 3.8E-01 | 4.9E-01 | -1.29 | 0.18917 |
| Apc2 | 4.3E-02 | 6.6E-02 | -1.54 | 0.13016 |
| Axin1 | 1.5E-02 | 2.4E-02 | -1.55 | 0.05900 |
| Axin2 | 2.5E-02 | 4.3E-02 | -1.76 | 0.00996 |
| Bcl9 | 3.3E-02 | 4.8E-02 | -1.46 | 0.17779 |
| Btrc | 2.3E-02 | 4.1E-02 | -1.75 | 0.02219 |
| Ctnnb1 | 2.3E-01 | 3.7E-01 | -1.63 | 0.08880 |
| Ccnd1 | 8.7E-02 | 1.4E-01 | -1.64 | 0.16283 |
| Ccnd2 | 9.1E-03 | 2.3E-02 | -2.49 | 0.03046 |
| Ccnd3 | 2.4E-02 | 4.4E-02 | -1.85 | 0.02811 |
| Csnk1a1 | 3.7E-01 | 4.9E-01 | -1.34 | 0.03945 |
| Csnk1d | 1.2E-01 | 1.8E-01 | -1.43 | 0.20033 |
| Csnk2a1 | 6.0E-02 | 9.0E-02 | -1.50 | 0.01109 |
| Csnk2b | 9.0E-01 | 1.2E+00 | -1.32 | 0.12559 |
| Daam1 | 2.4E-01 | 4.2E-01 | -1.76 | 0.04677 |
| Dixdc1 | 7.4E-02 | 1.2E-01 | -1.63 | 0.00247 |
| Dkk1 | 1.1E-03 | 1.2E-03 | -1.13 | 0.49486 |
| Dkk3 | 1.7E-01 | 2.7E-01 | -1.58 | 0.05506 |
| Dkk4 | 1.0E-01 | 1.7E-01 | -1.64 | 0.02177 |
| Dvl1 | 4.4E-02 | 6.8E-02 | -1.55 | 0.15926 |
| Dvl2 | 2.4E-02 | 3.0E-02 | -1.29 | 0.25291 |
| Dvl3 | 4.2E-02 | 7.1E-02 | -1.70 | 0.01509 |
| Ep300 | 6.3E-02 | 1.2E-01 | -1.96 | 0.00332 |
| Fbxw11 | 1.5E-01 | 2.1E-01 | -1.38 | 0.27901 |
| Fbxw2 | 6.7E-01 | 8.1E-01 | -1.20 | 0.29673 |
| Fgf4 | 5.6E-04 | 9.9E-04 | -1.78 | 0.05312 |
| Frzb | 5.9E-03 | 9.0E-03 | -1.53 | 0.26720 |
| Fzd1 | 1.4E-01 | 2.0E-01 | -1.49 | 0.12972 |
| Fzd2 | 1.9E-03 | 4.8E-03 | -2.47 | 0.06658 |
| Fzd3 | 1.4E-01 | 2.1E-01 | -1.55 | 0.02392 |
| Fzd4 | 7.4E-03 | 1.3E-02 | -1.71 | 0.03654 |
| Fzd5 | 2.2E-03 | 4.6E-03 | -2.08 | 0.16799 |
| Fzd6 | 7.0E-03 | 1.3E-02 | -1.91 | 0.00437 |
| Fzd7 | 2.2E-02 | 2.9E-02 | -1.28 | 0.20717 |
| Fzd9 | 2.0E-03 | 4.5E-03 | -2.26 | 0.09042 |
| Gsk3a | 4.8E-01 | 7.1E-01 | -1.47 | 0.07896 |
| Gsk3b | 5.6E-01 | 7.9E-01 | -1.41 | 0.07885 |
| Jun | 4.7E-02 | 6.5E-02 | -1.39 | 0.30477 |
| Kremen1 | 5.3E-03 | 9.0E-03 | -1.69 | 0.01538 |
| Lef1 | 1.5E-02 | 2.2E-02 | -1.52 | 0.07159 |
| Lrp5 | 3.2E-03 | 7.3E-03 | -2.24 | 0.00549 |
| Lrp6 | 1.0E-01 | 1.3E-01 | -1.26 | 0.17383 |
| Mitf | 2.0E-02 | 4.9E-02 | -2.45 | 0.13361 |
| Myc | 2.0E-03 | 4.0E-03 | -1.98 | 0.00666 |
| Nkd1 | 1.2E-02 | 2.6E-02 | -2.09 | 0.21437 |
| Nkd2 | 6.8E-03 | 1.3E-02 | -1.85 | 0.04517 |
| RGD1561440 | 2.4E-02 | 4.2E-02 | -1.80 | 0.00311 |
| Pitx2 | 1.7E-03 | 1.7E-03 | 1.03 | 0.88505 |
| Porcn | 2.3E-01 | 2.8E-01 | -1.24 | 0.16598 |
| Ppp2ca | 1.3E+00 | 1.8E+00 | -1.38 | 0.12881 |
| Ppp2r1a | 5.7E-01 | 9.0E-01 | -1.57 | 0.00621 |
| Pygo2 | 2.9E-03 | 6.3E-03 | -2.17 | 0.02359 |
| Rhoa | 1.2E+00 | 2.1E+00 | -1.71 | 0.00353 |
| Senp2 | 1.6E-01 | 2.6E-01 | -1.56 | 0.02894 |
| Sfrp1 | 2.3E-02 | 5.3E-02 | -2.30 | 0.02068 |
| Sfrp2 | 3.7E-03 | 7.7E-03 | -2.09 | 0.19428 |
| Sfrp4 | 8.2E-04 | 1.3E-03 | -1.56 | 0.00836 |
| Sfrp5 | 1.1E-03 | 1.6E-03 | -1.51 | 0.05005 |
| Tcf3 | 9.0E-03 | 1.7E-02 | -1.94 | 0.00054 |
| Tcf4 | 5.8E-01 | 7.8E-01 | -1.33 | 0.06372 |
| Tcf7 | 2.3E-03 | 3.3E-03 | -1.43 | 0.25535 |
| Tcfe2a | 8.1E-03 | 1.3E-02 | -1.59 | 0.14644 |
| Tle1 | 5.8E-02 | 9.1E-02 | -1.55 | 0.16625 |
| Tle2 | 4.1E-01 | 5.6E-01 | -1.39 | 0.34059 |
| Wif1 | 2.8E-03 | 5.0E-03 | -1.79 | 0.01399 |
| Wisp1 | 3.3E-03 | 4.5E-03 | -1.34 | 0.25818 |
| Wnt1 | 4.7E-04 | 5.5E-04 | -1.16 | 0.44400 |
| Wnt10a | 4.3E-04 | 4.8E-04 | -1.12 | 0.67497 |
| Wnt10b | 6.0E-04 | 6.3E-04 | -1.04 | 0.73714 |
| Wnt11 | 5.6E-03 | 1.2E-02 | -2.20 | 0.11090 |
| Wnt2 | 1.0E-03 | 1.4E-03 | -1.43 | 0.10151 |
| Wnt2b | 1.4E-02 | 2.1E-02 | -1.44 | 0.02325 |
| Wnt3 | 8.4E-03 | 1.3E-02 | -1.56 | 0.09550 |
| Wnt3a | 5.7E-04 | 8.2E-04 | -1.42 | 0.51981 |
| Wnt4 | 5.0E-03 | 1.1E-02 | -2.09 | 0.04808 |
| Wnt5a | 6.8E-03 | 1.8E-02 | -2.62 | 0.00029 |
| Wnt5b | 4.4E-03 | 1.0E-02 | -2.36 | 0.00360 |
| Wnt6 | 2.4E-03 | 3.0E-03 | -1.27 | 0.62605 |
| Wnt7a | 7.7E-03 | 1.5E-02 | -1.99 | 0.01008 |
| Wnt7b | 2.4E-03 | 3.8E-03 | -1.60 | 0.37925 |
| Wnt8a | 1.6E-03 | 1.7E-03 | -1.04 | 0.87043 |
| Wnt8b | 4.9E-03 | 7.4E-03 | -1.51 | 0.17107 |
| Wnt9a | 1.4E-03 | 2.4E-03 | -1.73 | 0.01723 |
| Wnt9b | 8.0E-04 | 1.4E-03 | -1.73 | 0.52746 |
Gene expression was measured in cerebellar tissue of P35 rats in which the pregnant dams were chronically fed with isocaloric ethanol-containing (37% caloric content) or
control (0% ethanol) liquid diets from gestation day 6 through delivery. Offspring were sacrificed and cerebella were harvested on P10
(not shown) or P35. Gene expression was measured in a targeted Wnt Pathway array that included 84 genes. (See gene full names, related Wnt pathways, and functions in
Table 1). Gene expression was calculated using the DDCt method with results normalized to HPRT (which did not change with ethanol exposure or age). Fold differences
were calculated. Data from 3 sets of pooled samples (3 cerebella each; 9 total per group) were analyzed using T-tests with corrections for multiple comparisons. Values in
Red indicate significant differences (P<0.05). Values in Blue reflect statistical trends (0.05<P<0.10).
Table 1: Wnt Targeted Array Results Comparing Gene Expression Levels in P35.
In P10 cerebella, we observed significant ethanol-associated foldincreases in Fgf4 (1.95), Wnt1 (2.29), and Wnt8a (2.01) expression. Otherwise, the gene expression levels were similar in the control and ethanol-exposed groups (data not shown). In contrast, in P35 rats, Wnt pathway gene expression was broadly inhibited by prenatal ethanol exposure. Ethanol significantly reduced (at least P<0.05) cerebellar expression of Axin2, Btrc, Cyclin D2 and D3, Casein kinase 2-alpha, DIX domain containing 1, DIckkopf homolog 4, Dishevelled 3, Dishevelled associated activator 1, E1A binding protein p300, Kringle containing transmembrane protein 1, low density lipoprotein receptorrelated protein 5 (LRP5), Myelocytomatosis oncogene (Myc), Naked cuticle homolog 2, regulatory protein phosphatase 2, Pygopus 2, Ras homolog gene family member A (RhoA), SUMO1, Secreted frizzledrelated proteins 1 (Sfrp1), Sfrp4, and Sfrp5, Frizzled homolog 3 (Fzd3), Fzd4, and Fzd6, Wnt inhibitory factor 1 (Wif1), Wnt 2b, Wnt4, Wnt5a, Wnt5b, Wnt7a, Wnt9a, and Transcription factor 3 (Tcf3) (Table 1). Ethanol inhibited expression of genes that mediate each of the three Wnt pathways, and regulate diverse functions such as cell growth, migration, survival, differentiation and morphogenesis.
Prenatal ethanol exposure differentially alters Wnt signaling in perinatal, juvenile and young adolescent brains
From the list of genes in which inter-group differences were 2-fold or greater in P35 cerebella, we extended our investigations to characterize effects of moderate and high doses of prenatal ethanol exposure on Wnt pathway gene expression at P3, P10, and P20. The objective was to determine how chronic gestational exposure to ethanol modulates Wnt signaling in the brain during the postnatal periods of active neuronal proliferation and migration (P3), cerebellar growth and myelination (P10), and dendritic arborization needed for motor learning and coordination (P20). For those studies, we used duplex qRT-PCR analysis to measure relative expression of: Wnt5a, Wnt5b, Frizzled (Fzd) 4, Fzd6, Axin2, Didxc, and EP300. Eight to 10 cerebella per group (different litters) were used in the reactions.
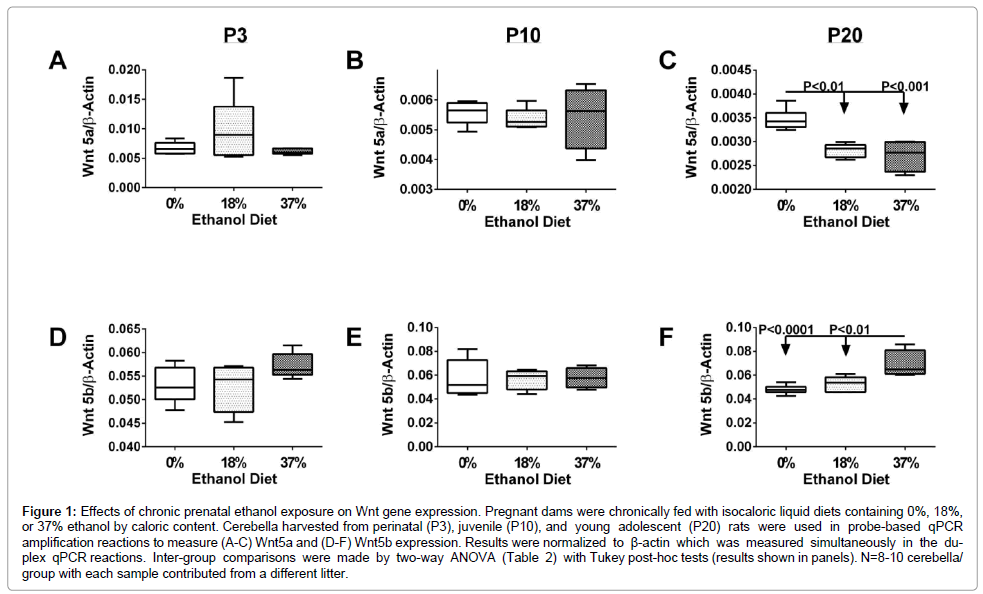
Wnt5a: Wnt5a signals through the Wnt/Ca2+ pathway and a ligand for Fzd5 and the tyrosine kinase orphan receptor 2. Wnt5a has an essential role in regulating developmental pathways during embryogenesis. Two-way ANOVA tests demonstrated significant effects of age (P<0.0001), and a trend (P=0.07) for interactions between age and ethanol dose on Wnt5a expression (Table 2). Wnt5a expression was similar for P3 and P10 control and ethanol-exposed cerebella. However, in P20 ethanol exposed (18% and 37%) cerebella, Wnt5a expression was significantly reduced relative to age-matched controls (P<0.01; P<0.001), and the P3 and P10 control and ethanol exposed cerebella (P<0.01) (Figures 1A-1C).
| Gene | Age | Ethanol | Age x Ethanol | |||
|---|---|---|---|---|---|---|
| F-Ratio | P-Value | F-Ratio | P-Value | F-Ratio | P-Value | |
| Wnt 5a | 22.79 | < 0.0001 | 1.66 | 0.2045 | 2.36 | 0.0706 |
| Wnt 5b | 2.128 | 0.5427 | 17.22 | 0.0116 | 16.53 | 0.0658 |
| Fzd4 | 157.82 | < 0.0001 | 1.59 | 0.2163 | 10.85 | < 0.0001 |
| Fzd6 | 2.78 | 0.0747 | 0.79 | 0.7530 | 1.97 | 0.1193 |
| Axin2 | 54.25 | < 0.0001 | 3.49 | 0.0390 | 9.19 | < 0.0001 |
| Didxc | 7.3 | 0.0021 | 1.15 | 0.3279 | 2.34 | 0.0727 |
| EP300 | 69.87 | < 0.0001 | 2.96 | 0.0632 | 4.46 | 0.0045 |
Quantitative duplex probe-based RT-PCR analysis was used to measure gene expression.Results were normalized to β-actin levels which remained stable with respect to ethanol treatment and postnatal age. Results were analyzed by Two-way ANOVA with post-hoc Tukey tests for significance. F-ratios and corresponding P-values for postnatal age, ethanol treatment and dose, and interaction between age and ethanol exposure are listed. Significant trends correspond to 0.05<P<0.10.
Table 2: Effects of Age and Ethanol Dose on Wnt Pathway Gene Expression.
Figure 1: Effects of chronic prenatal ethanol exposure on Wnt gene expression. Pregnant dams were chronically fed with isocaloric liquid diets containing 0%, 18%, or 37% ethanol by caloric content. Cerebella harvested from perinatal (P3), juvenile (P10), and young adolescent (P20) rats were used in probe-based qPCR amplification reactions to measure (A-C) Wnt5a and (D-F) Wnt5b expression. Results were normalized to β-actin which was measured simultaneously in the duplex qPCR reactions. Inter-group comparisons were made by two-way ANOVA (Table 2) with Tukey post-hoc tests (results shown in panels). N=8-10 cerebella/ group with each sample contributed from a different litter.
Wnt5b: Wnt5b ligand signals through the Wnt/Ca2+ pathway in the regulation of several developmental processes, as well as oncogenesis. Two-way ANOVA tests demonstrated significant effects of ethanol dose (P=0.0116), and a trend for age x ethanol dose interactions (P=0.0658) on Wnt5b expression (Table 2). Wnt5b expression was similar in P3, P10, and P20 control and 18% ethanol-exposed cerebella. In contrast, at P3 and P20, prenatal exposure to the 37% ethanol diet resulted in higherlevels of Wnt5b relative to the corresponding control and 18% ethanol diet groups, although the differences reached significance only at P20 (P<0.05) (Figures 1D-1F). Age x ethanol dose effects were manifested by significant further increases in Wnt5b expression in P20- 37% ethanol versus P3-37% ethanol (P<0.05), and the P3 (P<0.01) and P10 (P<0.05) 18% ethanol diets. Similarly, in a previous study it was found that Wnt5b expression increased with brain maturation [55].
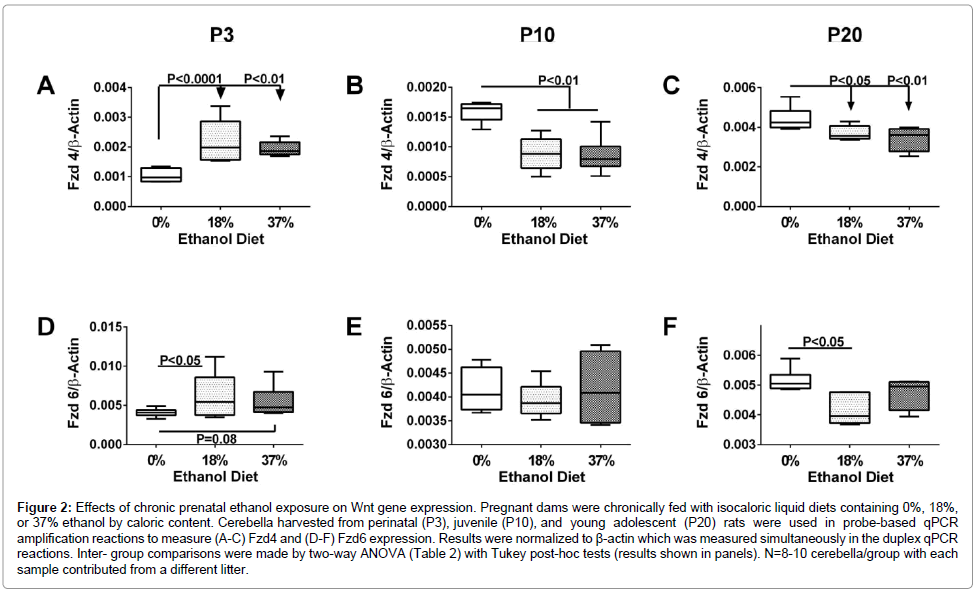
Fzd4: Fzd4 receptor participates in the canonical pathway and promotes Wnt signaling by activating Dishevelled which inhibits GSK-3β. Fzd4 gene expression was significantly modulated with age and ethanol dose (Figure 2), and correspondingly, two-way ANOVA tests demonstrated significant effects of age (P<0.0001) and age x ethanol dose (P<0.0001) on Fzd4 mRNA levels. Among controls, Fzd4 expression increased with age (brain maturation), particularly between P10 and P20 (P<0.0001), consistent with a previous report [55]. Fzd4 expression also increased with age in ethanol-exposed cerebella, but the responses were less pronounced than in the control group. The effects of ethanol on the relative levels in Fzd gene expression were more related to age than ethanol dose. At P3, Fzd4 expression was increased in ethanol-exposed relative to control cerebella (Figure 2A, 2B), whereas at P10 and P20 Fzd4 expression was reduced. Effects of ethanol were not dose-related at P3 or P10, but at P20, Fzd4 expression declined with ethanol dose (Figure 2C).
Figure 2: Effects of chronic prenatal ethanol exposure on Wnt gene expression. Pregnant dams were chronically fed with isocaloric liquid diets containing 0%, 18%, or 37% ethanol by caloric content. Cerebella harvested from perinatal (P3), juvenile (P10), and young adolescent (P20) rats were used in probe-based qPCR amplification reactions to measure (A-C) Fzd4 and (D-F) Fzd6 expression. Results were normalized to β-actin which was measured simultaneously in the duplex qPCR reactions. Inter- group comparisons were made by two-way ANOVA (Table 2) with Tukey post-hoc tests (results shown in panels). N=8-10 cerebella/group with each sample contributed from a different litter.
Fzd6: Fzd6 receptor signals through the canonical pathway and mediates tissue polarity during development. In contrast to Fzd4, Fzd6 mRNA levels were modestly modulated across the three age groups and with respect to ethanol exposure (Figures 2D–2F). Two-way ANOVA tests revealed a trend for age effects (P=0.075), but not for ethanol dose or ethanol’s interaction with age (Table 2). However, in P20 control cerebella, Fzd6 expression was sharply increased relative to all P3 and P10, and the ethanol-exposed P20 groups (P<0.05). Post tests revealed higher levels of Fzd6 in the 18% (P<0.05) and 37% (P=0.085; trend) ethanol groups relative to control at P3, and lower levels of Fzd6 in P20 ethanol-exposed relative to corresponding control cerebella (Figure 3). At P10, Fzd6 expression did not significantly vary with ethanol treatment or dose.
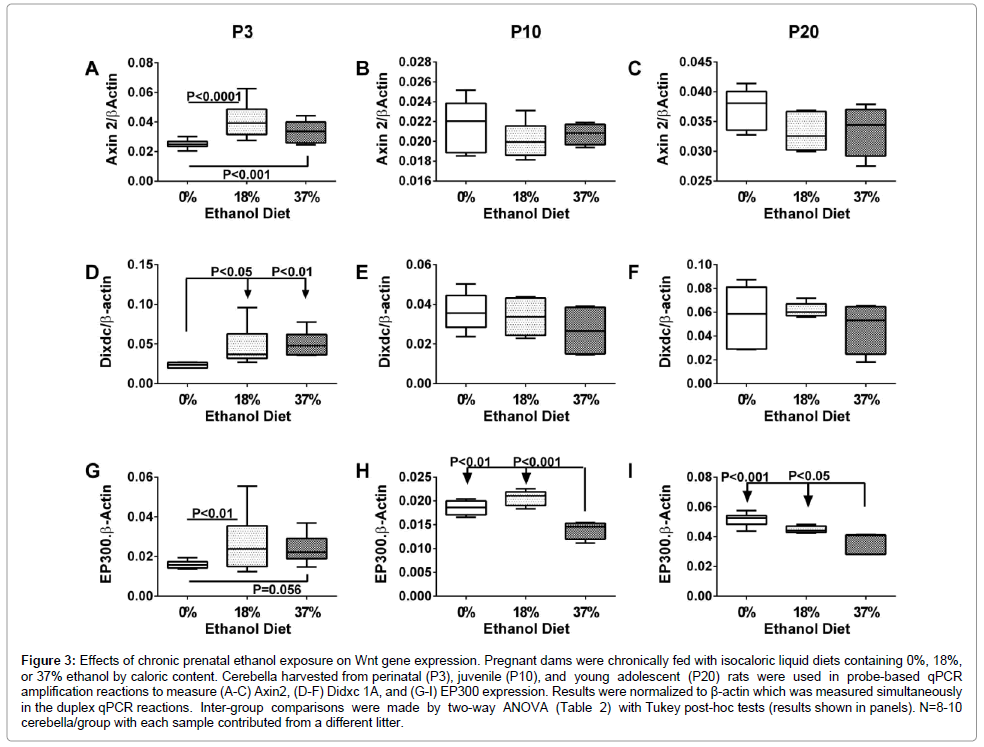
Figure 3: Effects of chronic prenatal ethanol exposure on Wnt gene expression. Pregnant dams were chronically fed with isocaloric liquid diets containing 0%, 18%, or 37% ethanol by caloric content. Cerebella harvested from perinatal (P3), juvenile (P10), and young adolescent (P20) rats were used in probe-based qPCR amplification reactions to measure (A-C) Axin2, (D-F) Didxc 1A, and (G-I) EP300 expression. Results were normalized to β-actin which was measured simultaneously in the duplex qPCR reactions. Inter-group comparisons were made by two-way ANOVA (Table 2) with Tukey post-hoc tests (results shown in panels). N=8-10 cerebella/group with each sample contributed from a different litter.
Axin2: Axin2 is involved in the regulation of canonical Wnt signaling and participates in the destruction complex to stabilize β-catenin, causing it to accumulate and eventually translocate to the nucleus. Developmental age and ethanol exposure effects on Axin2 mimicked those of Fzd4. In particular, moderate but significant increases in Axin2 expression occurred in all groups between P3 and P10 (P<0.01 to P<0.0001), whereas between P3 or P10 and P20, Axin2 expression was increased sharply in all groups (P<0.0001; Figures 3A-3C). Correspondingly, two-way ANOVA tests demonstrated significant effects of postnatal age (P<0.0001), ethanol dose (P=0.039), and interactions between age and ethanol dose (P<0.0001) on Axin2 expression (Table 2). Post hoc significance tests further demonstrated that at P3, Axin2 expression was significantly higher in the 18% (P<0.0001) and 37% (P<0.001) ethanol groups relative to control, whereas at P10 and P20, there were no significant differences caused by ethanol exposure.
Dixdc: The DIX domain-containing 1 gene is a positive regulator of canonical Wnt signaling, and functions by interacting with Axin1, Dishevelled 2, and MAP3K1. In control cerebella, Dixdc expression increased stepwise with age from P3 to P20. Ethanol-exposed cerebella showed similar trends, but the responses were smaller. Twoway ANOVA tests demonstrated significant age-dependent effects (P=0.0021) and a trend for age x ethanol dose interactive effects (P=0.073) on Dixdc expression (Table 2). Graphically, at P3, ethanol exposure increased Dixdc in the 18% (P<0.05) and 37% (P<0.01) ethanol diet groups. However, at P10 and P20, the trends regarding ethanol’s effects were neutralized resulting in similar levels of Dixdc in control and ethanol diet groups (Figures 3D-3F).
EP300: E1A binding protein p300 regulates cell growth, division, and differentiation via canonical Wnt signaling. EP300 functions as a transcriptional co-activator and histone acetyltransferase, regulating transcription via chromatin remodeling. EP300 expression was unchanged between P3 and P10, but the levels increased sharply between P10 and P20, similar to Fzd4, Axin2, and Dixdc. Although the ethanol-exposed groups manifested similar responses, the agedependent trends were less robust than control. Two-way ANOVA tests demonstrated significant effects of developmental age (P<0.0001) and age x ethanol dose interactions (P=0.0045), and a trend for ethanol dose effects (P=0.063) on EP300 expression (Table 2). At P3, EP300 expression was higher in the 18% (P<0.01) and 37% (P=0.056) ethanolexposed cerebella relative to controls. At P10, ethanol exposure and dose did not significantly modulate cerebellar EP300 expression. At P20, EP300 expression was significantly reduced in the 37% ethanol diet group relative to control (P<0.001). EP300 was also lower in the 18% ethanol diet group, but the difference from control did not reach significance (P=0.11) (Figures 3G-3I).
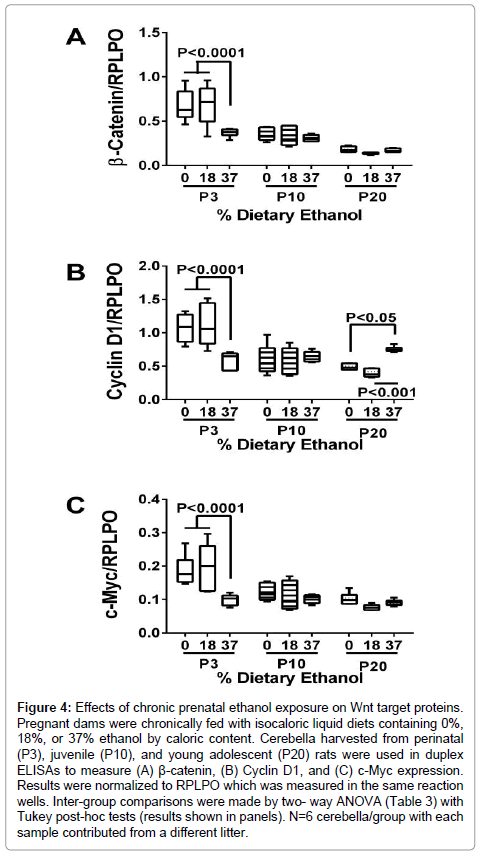
Chronic gestational exposure to ethanol differentially alters Wnt target protein expression in perinatal, juvenile and young adolescent cerebella
The studies were extended to examine effects of ethanol on Wnt target protein expression at P3, P10, and P20. For those studies, we used duplex ELISAs to measure relative expression of: β-catenin, Cyclin D1, and C-Myc. Six cerebella per group (different litters) were used in the reactions, and all assays were performed in quadruplicate. Two-way ANOVA tests demonstrated significant effects of age (P<0.0001) and interactions between age and ethanol exposure (at least P=0.001) with respect to all three proteins, and significant effects of ethanol dose in relation to β-catenin (P=0.005) and C-Myc (P=0.003) expression (Table 3). The graphed results illustrate three major trends: 1) cerebellar protein expression of β-Catenin, Cyclin D1, and c-Myc decline with age; and 2) at P3, all three proteins were significantly reduced by exposure to the 37% but not the 18% ethanol-containing diet; and 3) the inter-group differences at P10 and P20 were modest except for the significantly higher levels of Cyclin D1 measured in the P20 37% ethanol-diet group relative to control and the 18% ethanol diet group (Figure 4).
| Protein | Age | Ethanol | Age x Ethanol | |||
|---|---|---|---|---|---|---|
| F-Ratio | P-Value | F-Ratio | P-Value | F-Ratio | P-Value | |
| β-Catenin | 66.65 | < 0.0001 | 5.95 | 0.0051 | 5.39 | 0.0012 |
| Cyclin D1 | 25.78 | < 0.0001 | 0.56 | N.S. | 11.93 | <0.00018 |
| c-Myc | 21.48 | < 0.0001 | 6.67 | 0.0029 | 5.10 | 0.0018 |
Duplex ELISAs were used to measure immunoreactivity. Results were normalized to large ribonuclear protein which remained stable with respect to ethanol treatment and postnatal age. Results were analyzed by Two-way ANOVA with post-hoc Tukey tests for significance. F-ratios and corresponding P-values for postnatal age, ethanol treatment and dose, and interaction between age and ethanol exposure are listed. N.S.= Not Significant
Table 3: Effects of Age and Ethanol Dose on Wnt Target Protein Expression.
Figure 4: Effects of chronic prenatal ethanol exposure on Wnt target proteins. Pregnant dams were chronically fed with isocaloric liquid diets containing 0%, 18%, or 37% ethanol by caloric content. Cerebella harvested from perinatal (P3), juvenile (P10), and young adolescent (P20) rats were used in duplex ELISAs to measure (A) β-catenin, (B) Cyclin D1, and (C) c-Myc expression. Results were normalized to RPLPO which was measured in the same reaction wells. Inter-group comparisons were made by two- way ANOVA (Table 3) with Tukey post-hoc tests (results shown in panels). N=6 cerebella/group with each sample contributed from a different litter.
Discussion
Potential role of impaired Wnt signaling in FASD
In previous studies using the same experimental models, we showed that chronic prenatal ethanol exposures cause cerebellar hypoplasia with reductions in granule and Purkinje cell populations, hypofoliation (simplified folding patterns), and disordered neuronal migration [15,53,56] with sustained impairments in motor function [57,58]. Ethanol-mediated reductions in the neuronal populations are likely due to combined effects of decreased proliferation and survival, mediated in part by deficits in insulin, IGF [15,16,59-61], and neurotrophin signaling [62-65]. However, growth in our understanding of how disruption of insulin/IGF networks contribute to the teratogenic effects of ethanol, led to further questions regarding the roles of other signaling pathways, particularly those that cross-talk with insulin/IGF networks to modulate development and neuronal functions. Indeed, there is evidence that IGF-1 receptor signaling can cross-talk with the canonical Wnt pathway via inhibition of GSK- 3β and attendant increased levels of β-catenin, leading to enhanced cellular proliferation/neurogenesis in the brain [66]. Therefore, in addition to impaired neuronal survival [53,56], cerebellar hypoplasia in FASD could be mediated by inhibition of cell growth via decreased signaling through the IGF-1 receptor, Wnt/β-catenin, or cross-talk between IGF-1 receptor and canonical Wnt.
Mechanistically, ethanol inhibition of insulin/IGF signaling during developmental increases GSK-3β activity in the brain [15,47,48,67]. High levels of GSK-3β promote oxidative stress, DNA damage, and mitochondrial dysfunction, and inhibit neuronal survival and migration [47,67-70]. In FASD, increased levels of GSK-3β persist in adolescent cerebella [15,71-75]. GSK-3β inhibits Wnt signaling due to phosphorylation and proteasomal degradation of β-catenin [76]. Consequently, Wnt target genes that mediate various functions including growth, survival, and plasticity, fail to be stimulated. Since Wnt pathways cross-talk with insulin/IGF [66], and both have critical roles in brain development and function, future studies could further assess the interdependence of these networks on cerebellar development by immune depletion or targeted inhibition of IGFs [77], or the restoration of insulin/IGF-1 functions in FASD models by treatment with insulin sensitizer agents [3,78]. Herein, we conducted experiments to examine the nature and degree of cerebellar Wnt signaling was dysregulated in our FASD model. To date, very limited information is available about ethanol’s effects on Wnt signaling in postnatal developing and adolescent brains. The significance of this work is that the results suggest additional targets for therapeutic rescue of neurodevelopmental defects in FASD.
Broad disruption of Wnt signaling in FASD
We used a targeted array to assess the extent of Wnt signaling abnormalities in cerebella of P10 and P35 rats that had been subjected to chronic prenatal ethanol exposure (37% ethanol diet by caloric content; 9.2% v/v). Those time points were selected because they correspond with the active (P10) and completed (P35) stages of cerebellar development. Targeted array approaches enable simultaneous interrogation of genes that regulate different components of Wnt signaling, including brain morphogenesis, neuronal proliferation, migration, differentiation, and axon outgrowth (Supplementary Table 1).
The main findings were that chronic prenatal ethanol exposure significantly altered Wnt pathway gene expression, but the effects differed with respect to postnatal developmental stage. Only a few intergroup differences were observed at P10, and all were associated with relative increases in Wnt or Wnt target gene expression. In contrast, at P35, the consequences of prenatal ethanol exposure were broad and associated with significantly reduced expression of several Wnt ligands and Frizzled receptors, components of the destruction complex, and targets that mediate neuronal growth, survival, differentiation, and plasticity, corresponding with previous observations in other models of FASD [41,79]. Although most of the affected genes pertained to canonical pathway, genes within the Wnt/Calcium and Planar/Polar pathways were also inhibited (Table 1). Therefore, in young adolescent cerebella in which neurodevelopment was largely completed, Wnt signaling mechanisms were substantially impaired by chronic prenatal ethanol exposure. Therefore, the consequences of chronic prenatal ethanol exposure may shift over time in relation to postnatal developmental and maturation of the brain.
Effects of prenatal ethanol exposure on Wnt signaling at different stages of postnatal cerebellar development
The array data prompted us to further examine the expression of selected Wnt pathway genes at different postnatal ages, and following chronic prenatal exposure to moderate (18% diet) or high (37% diet) levels of ethanol. Dose effects were of interest because severity of the phenotype and molecular signaling abnormalities increase with levels of prenatal ethanol exposure [15,53]. The shifts in direction and magnitude of ethanol’s effects on Wnt pathway genes with age and brain maturation were illuminating and generally supportive of the targeted array results. In P3 cerebella in which neurogenesis is prominent, Wnt pathway gene expression was mainly up-regulated following ethanol exposure, whereas at P10, differences from control were either modestly inhibitory or non-existent, and at P35, the effects of ethanol were broadly inhibitory. Dose effects were observed for only some genes. These dynamic shifts in postnatal responses to prenatal ethanol exposures curiously hint at fetal programing [80,81], and suggest that the consequences of prenatal ethanol exposure may go undetected if the proper tools and time points are not used to assess their effects.
In P3 cerebella in which the inhibitory effects of ethanol on neuronal survival and proliferation are prominent and correlate with impairments in insulin/IGF signaling through Akt [53,56,59], Wnt signaling mechanisms were found to be up-regulated. Conceivably, the increased expression of Wnt pathway genes may reflect a compensatory response to deficits in insulin/IGF signaling. Previous studies showed that ethanol-induced impairments in cerebellar insulin/IGF signaling are sustained through early adolescence [58,82]. However, in contrast to the findings at P3, Wnt gene expression was relatively unaltered at P10, and broadly suppressed at P20 and P35. These observations correspond with previous work showing that the effects of early developmental exposures to ethanol on Wnt7a and Cyclin D2 expression are modulated between P0 and P15 when the cerebellar cortical neurons transition from a relative alcohol-vulnerable to alcohol-resistant state [41]. In addition, another report showed that low or high levels of ethanol exposure (in vitro) inhibits Wnt3a, Wnt 5a, LRP6, DVL2 and β-catenin in differentiation neuronal cells [42]. We hypothesize that the shifts from possible compensatory increases in Wnt at P3, to minimal responses at P10, and inhibition at P20 and P35 reflect progressive uncoupling of the cross-talk between insulin/IGF and Wnt networks. Developmentally, before P9, cerebellar Purkinje neurons are immature, from P10-P18, they undergo rapid maturation, and from P20 and beyond, they are essentially mature [83-86]. Between P10 and P35, cerebellar myelination [87] and neuronal plasticity [83] are robust, and both are regulated by insulin/IGF and Wnt signaling [17,43,82,88-94]. Therefore, reduced white matter integrity and volume and deficits in neuronal plasticity, which are well-recognized long-term CNS abnormalities in FASD [13,95-100], could be mediated by dual inhibition of insulin/IGF and Wnt in the brain. The probable contribution of both pathways is supported by the finding that treatment with insulin sensitizer agents failed to completely restore cerebellar structure and gene expression in our FASD model [82]. On the other hand, inhibition of Wnt gene expression could represent an important mediator of brain insulin resistance since Wnt/β-catenin signaling increases cellular sensitivity to insulin [101]
Prenatal ethanol exposure inhibited expression of several Fzd and Wnt gene isoforms, together with multiple genes that participate in the destruction complex. Therefore, the long-term effects of chronic prenatal ethanol exposure include broad inhibition of Wnt signaling at multiple levels within the cascades and through all major pathways. In contrast to previous studies in which we showed that the adverse effects of chronic prenatal ethanol exposures were largely dose-dependent, herein, we observed relatively modest ethanol dose-related effects on Wnt signaling and prominent effects of postnatal age. This suggests that ethanol’s effects on Wnt signaling and gene regulation may be thresholded, and beyond a certain level of exposure, responses may be nearly all-or-none. The degree to which prenatal alcohol exposures differentially modulate insulin/IGF, Wnt, and probably other pathways at different stages of development could help account for the phenotypic variability in FASD. Further studies are needed to characterize specific and independent roles of impaired Wnt versus insulin/IGF signaling in FASD, and better define therapeutic strategies needed to reduce longterm adverse effects of prenatal alcohol exposures.
References
- Riley EP, Infante MA, Warren KR (2011) Fetal alcohol spectrum disorders: an overview. Neuropsychol Rev 21: 73-80.
- Mattson SN, Crocker N, Nguyen TT (2011) Fetal alcohol spectrum disorders: neuropsychological and behavioral features. Neuropsychol Rev 21: 81-101.
- De la Monte S, J.R Wands (2010) Role of central nervous system insulin resistance in fetal alcohol spectrum disorders. Journal of population therapeutics and clinical pharmacology Journal de la therapeutique des populations et de la pharamcologie clinique. 17: e390-404.
- de la Monte SM, Wands JR (2005) Review of insulin and insulin-like growth factor expression, signaling, and malfunction in the central nervous system: relevance to Alzheimer's disease. J Alzheimers Dis 7: 45-61.
- Chesik D, De Keyser J, Wilczak N (2008) Insulin-like growth factor system regulates oligodendroglial cell behavior: therapeutic potential in CNS. J Mol Neurosci 35: 81-90.
- Gong X, Xie Z, Zuo H (2008) Invivo insulin deficiency as a potential etiology for demyelinating disease. Med Hypotheses 71: 399-403.
- Liang G, Cline GW, Macica CM (2007) IGF-1 stimulates de novo fatty acid biosynthesis by Schwann cells during myelination. Glia 55: 632-641.
- Ye P, Kollias G, D'Ercole AJ (2007) Insulin-like growth factor-I ameliorates demyelination induced by tumor necrosis factor-alpha in transgenic mice. J Neurosci Res 85: 712-722.
- Laviola L, Natalicchio A, Perrini S, Giorgino F (2008) Abnormalities of IGF-I signaling in the pathogenesis of diseases of the bone, brain, and fetoplacental unit in humans. Am J Physiol Endocrinol Metab 295: E991-999.
- Broughton SK, Chen H, Riddle A, Kuhn SE, Nagalla S, et al. (2007) Large-scale generation of highly enriched neural stem-cell-derived oligodendroglial cultures: maturation-dependent differences in insulin-like growth factor-mediated signal transduction. J Neurochem. 100: 628-638.
- Joseph D'Ercole A, Ye P (2008) Expanding the mind: insulin-like growth factor I and brain development. Endocrinology 149: 5958-5962.
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, et al. (1997) Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 275: 661-665.
- Wozniak JR, Muetzel RL (2011) What does diffusion tensor imaging reveal about the brain and cognition in fetal alcohol spectrum disorders? Neuropsychol Rev 21: 133-147.
- de la Monte SM, Neely TR, Cannon J, Wands JR (2001) Ethanol impairs insulin-stimulated mitochondrial function in cerebellar granule neurons. Cell Mol Life Sci 58: 1950-1960.
- de la Monte SM, Tong M, Carlson RI, Carter JJ, Longato L, et al. (2009) Ethanol inhibition of aspartyl-asparaginyl-beta-hydroxylase in fetal alcohol spectrum disorder: potential link to the impairments in central nervous system neuronal migration. Alcohol 43: 225-240.
- de la Monte SM, Tong M, Cohen AC, Sheedy D, Harper C, et al. (2008) Insulin and insulin-like growth factor resistance in alcoholic neurodegeneration. Alcohol Clin Exp Res 32: 1630-1644.
- Freude S, Leeser U, Müller M, Hettich MM, Udelhoven M, et al. (2008) IRS-2 branch of IGF-1 receptor signaling is essential for appropriate timing of myelination. J Neurochem 107: 907-917.
- Singh AK, Gupta S, Jiang Y, Younus M, Ramzan M (2009) In vitro neurogenesis from neural progenitor cells isolated from the hippocampus region of the brain of adult rats exposed to ethanol during early development through their alcohol-drinking mothers. Alcohol Alcohol. 44: 185-98.
- Yi F, Sun J, Lim GE, Fantus IG, Brubaker PL, et al. (2008) Cross talk between the insulin and Wnt signaling pathways: evidence from intestinal endocrine L cells. Endocrinology 149: 2341-2351.
- Beildeck ME, Gelmann EP, Byers SW (2010) Cross-regulation of signaling pathways: an example of nuclear hormone receptors and the canonical Wnt pathway. Exp Cell Res 316: 1763-1772.
- Cadigan KM, Peifer M (2009) Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol 1: a002881.
- Sugimura R, Li L (2010) Noncanonical Wnt signaling in vertebrate development, stem cells, and diseases. Birth Defects Res C Embryo Today 90: 243-256.
- Rao Q, Liu XH, Zhou HB, Ma HH, Lu ZF, et al. (2010) Expression analysis of Wnt-5a in renal epithelial neoplasms: distinguishing renal oncocytoma from a wide spectrum of renal cell carcinomas. Tumori 96: 304-309.
- Tang SJ (2007) The synaptic Wnt signaling hypothesis. Synapse 61: 866-868.
- Chen J, Park CS, Tang SJ (2006) Activity-dependent synaptic Wnt release regulates hippocampal long term potentiation. J Biol Chem 281: 11910-11916.
- Miller JR, Hocking AM, Brown JD, Moon RT (1999) Mechanism and function of signal transduction by the Wnt/beta-catenin and Wnt/Ca2+ pathways. Oncogene 18: 7860-7872.
- Shulman JM, Perrimon N, Axelrod JD (1998) Frizzled signaling and the developmental control of cell polarity. Trends Genet 14: 452-458.
- Wansleeben C, Meijlink F (2011) The planar cell polarity pathway in vertebrate development. Dev Dyn 240: 616-626.
- LaMonica K, Bass M, Grabel L (2009) The planar cell polarity pathway directs parietal endoderm migration. Dev Biol 330: 44-53.
- James RG, Conrad WH, Moon RT (2008) Beta-catenin-independent Wnt pathways: signals, core proteins, and effectors. Methods Mol Biol 468: 131-144.
- Tanegashima K, Zhao H, Dawid IB (2008) WGEF activates Rho in the Wnt-PCP pathway and controls convergent extension in Xenopus gastrulation. EMBO J 27: 606-617.
- Liu H, Fergusson MM, Wu JJ, Rovira II, Liu J, et al. (2011) Wnt signaling regulates hepatic metabolism. Sci Signal 4: ra6.
- Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, et al. (2010) Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev 24: 1507-1518.
- Bommer GT, Feng Y, Iura A, Giordano TJ, Kuick R, et al. (2010) IRS1 regulation by Wnt/beta-catenin signaling and varied contribution of IRS1 to the neoplastic phenotype. J Biol Chem 285: 1928-1938.
- Yamazaki H, Yanagawa Si (2003) Axin and the Axin/Arrow-binding protein DCAP mediate glucose-glycogen metabolism. Biochem Biophys Res Commun 304: 229-235.
- Ding VW, Chen RH, McCormick F (2000) Differential regulation of glycogen synthase kinase 3beta by insulin and Wnt signaling. J Biol Chem 275: 32475-32481.
- Pearl LH, Barford D (2002) Regulation of protein kinases in insulin, growth factor and Wnt signalling. Curr Opin Struct Biol 12: 761-767.
- Wang LL, Yang AK, He SM, Liang J, Zhou ZW, et al. (2010) Identification of molecular targets associated with ethanol toxicity and implications in drug development. Curr Pharm Des 16: 1313-1355.
- Lauing KL, Roper PM, Nauer RK, Callaci JJ (2012) Acute alcohol exposure impairs fracture healing and deregulates β-catenin signaling in the fracture callus. Alcohol Clin Exp Res 36: 2095-2103.
- Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, et al. (2007) Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev Dyn 236: 613-631.
- Karacay B, Li S, Bonthius DJ (2008) Maturation-dependent alcohol resistance in the developing mouse: cerebellar neuronal loss and gene expression during alcohol-vulnerable and -resistant periods. Alcoholism, clinical and experimental research. 32: 1439-50.
- Vangipuram SD, Lyman WD (2012) Ethanol affects differentiation-related pathways and suppresses Wnt signaling protein expression in human neural stem cells. Alcohol Clin Exp Res 36: 788-797.
- Patapoutian A, Reichardt LF (2000) Roles of Wnt proteins in neural development and maintenance. Curr Opin Neurobiol 10: 392-399.
- Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R (2000) Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development 127: 469-482.
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S (1997) Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 389: 966-970.
- Salinas PC, Fletcher C, Copeland NG, Jenkins NA, Nusse R (1994) Maintenance of Wnt-3 expression in Purkinje cells of the mouse cerebellum depends on interactions with granule cells. Development 120: 1277-1286.
- de la Monte SM, Wands JR (2002) Chronic gestational exposure to ethanol impairs insulin-stimulated survival and mitochondrial function in cerebellar neurons. Cell Mol Life Sci. 59: 882-93.
- Carter JJ, Tong M, Silbermann E, Lahousse SA, Ding FF, et al. (2008) Ethanol impaired neuronal migration is associated with reduced aspartyl-asparaginyl-beta-hydroxylase expression. Acta Neuropathol 116: 303-315.
- Tanneberger K, Pfister AS, Kriz V, Bryja V, Schambony A, et al. (2011) Structural and functional characterization of the Wnt inhibitor APC membrane recruitment 1 (Amer1). J Biol Chem 286: 19204-19214.
- Roberts DM, Pronobis MI, Poulton JS, Waldmann JD, Stephenson EM, et al. (2011) Deconstructing the ßcatenin destruction complex: mechanistic roles for the tumor suppressor APC in regulating Wnt signaling. Mol Biol Cell 22: 1845-1863.
- MacDonald BT, Tamai K, He X (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9-26.
- Gundogan F, Elwood G, Longato L, Tong M, Feijoo A, et al. (2008) Impaired placentation in fetal alcohol syndrome. Placenta 29: 148-157.
- Soscia SJ, Tong M, Xu XJ, Cohen AC, Chu J, et al. (2006) Chronic gestational exposure to ethanol causes insulin and IGF resistance and impairs acetylcholine homeostasis in the brain. Cell Mol Life Sci 63: 2039-2056.
- Gundogan F, Elwood G, Greco D, Rubin LP, Pinar H, et al. (2007) Role of aspartyl-(asparaginyl) beta-hydroxylase in placental implantation: Relevance to early pregnancy loss. Hum Pathol 38: 50-59.
- Choi MR, Jung KH, Park JH, Das ND, Chung MK, et al. (2011) Ethanol-induced small heat shock protein genes in the differentiation of mouse embryonic neural stem cells. Arch Toxicol 85: 293-304.
- Xu J, Yeon JE, Chang H, Tison G, Chen GJ, et al. (2003) Ethanol impairs insulin-stimulated neuronal survival in the developing brain: role of PTEN phosphatase. J Biol Chem 278: 26929-26937.
- Goodlett CR, Thomas JD, West JR (1991) Long-term deficits in cerebellar growth and rotarod performance of rats following "binge-like" alcohol exposure during the neonatal brain growth spurt. Neurotoxicol Teratol 13: 69-74.
- Ewenczyk AE, Ziplow J, Tong M, Le T, de la Monte M (2012) Sustained impairments in brain insulin/IGF signaling in adolescent rats subjected to binge alcohol exposure during development. J Clin Exp Pathol. 2(2): 106.
- de la Monte SM, Xu XJ, Wands JR (2005) Ethanol inhibits insulin expression and actions in the developing brain. Cell Mol Life Sci 62: 1131-1145.
- Hallak H, Seiler AE, Green JS, Henderson A, Ross BN, et al. (2001) Inhibition of insulin-like growth factor-I signaling by ethanol in neuronal cells. Alcohol Clin Exp Res 25: 1058-1064.
- Zhang FX, Rubin R, Rooney TA (1998) Ethanol induces apoptosis in cerebellar granule neurons by inhibiting insulin-like growth factor 1 signaling. J Neurochem 71: 196-204.
- Heaton MB, Mitchell JJ, Paiva M (1999) Ethanol-induced alterations in neurotrophin expression in developing cerebellum: relationship to periods of temporal susceptibility. Alcohol Clin Exp Res 23: 1637-1642.
- Neveu I, Arenas E (1996) Neurotrophins promote the survival and development of neurons in the cerebellum of hypothyroid rats in vivo. J Cell Biol 133: 631-646.
- Seil FJ (1999) BDNF and NT-4, but not NT-3, promote development of inhibitory synapses in the absence of neuronal activity. Brain Res 818: 561-564.
- Light KE, Brown DP, Newton BW, Belcher SM, Kane CJ (2002) Ethanol-induced alterations of neurotrophin receptor expression on Purkinje cells in the neonatal rat cerebellum. Brain Res 924: 71-81.
- Hu Q, Lee SY, O'Kusky JR, Ye P (2012) Signaling Through the Type 1 Insulin-Like Growth Factor Receptor (IGF1R) Interacts with Canonical Wnt Signaling to Promote Neural Proliferation in Developing Brain. ASN Neuro .
- Luo J (2010) Lithium-mediated protection against ethanol neurotoxicity. Front Neurosci 4: 41.
- Mora A, Sabio G, Alonso JC, Soler G, Centeno F (2002) Different dependence of lithium and valproate on PI3K/PKB pathway. Bipolar Disord 4: 195-200.
- Hughes JP, Ward DR, Facci L, Richardson JC, Skaper SD (2010) Apoptosis-associated tyrosine kinase and neuronal cell death. Neurochem Res 35: 588-597.
- Pap M, Cooper GM (1998) Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-Kinase/Akt cell survival pathway. J Biol Chem 273: 19929-19932.
- Mooney SM, Miller MW (2003) Ethanol-induced neuronal death in organotypic cultures of rat cerebral cortex. Brain Res Dev Brain Res 147: 135-141.
- Lahousse SA, Carter JJ, Xu XJ, Wands JR, de la Monte SM (2006) Differential growth factor regulation of aspartyl-(asparaginyl)-beta-hydroxylase family genes in SH-Sy5y human neuroblastoma cells. BMC Cell Biol 7: 41.
- Lawton M, Tong M, Gundogan F, Wands JR, de la Monte SM (2010) Aspartyl-(asparaginyl) beta-hydroxylase, hypoxia-inducible factor-alpha and Notch cross-talk in regulating neuronal motility. Oxidative medicine and cellular longevity. 3: 347-56.
- Maeda T, Sepe P, Lahousse S, Tamaki S, Enjoji M, et al. (2003) Antisense oligodeoxynucleotides directed against aspartyl (asparaginyl) beta-hydroxylase suppress migration of cholangiocarcinoma cells. J Hepatol 38: 615-622.
- Silbermann E, Moskal P, Bowling N, Tong M, de la Monte SM (2010) Role of aspartyl-(asparaginyl)-β-hydroxylase mediated notch signaling in cerebellar development and function. Behav Brain Funct 6: 68.
- Doble BW, Woodgett JR (2003) GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci 116: 1175-1186.
- Agis-Balboa RC, Arcos-Diaz D, Wittnam J, Govindarajan N, Blom K, et al. (2011) A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J 30: 4071-4083.
- Le T, Tong M, Nguyen VA, Sde la Monte SM (2013) PPAR Agonist Rescue of Ethanol-Impaired Brain Insulin Signaling:Cerebellar Slice Culture Model. Journal of Drug and Alcohol Research. 2: 9.
- Flentke GR, Garic A, Amberger E, Hernandez M, Smith SM (2011) Calcium-mediated repression of beta-catenin and its transcriptional signaling mediates neural crest cell death in an avian model of fetal alcohol syndrome. Birth Defects Res A Clin Mol Teratol. 91(7): 591-602.
- Haycock PC (2009) Fetal alcohol spectrum disorders: the epigenetic perspective. Biol Reprod 81: 607-617.
- Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, et al. (2008) Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res 32: 1525-1534.
- Ewenczyk AE, Conoway A, Ziplow J, Le T, Q-G.L Nguyen, et al. (2013) PPAR-d agonist rescue of long-term neurobehavioral and brain insulin/IGF signaling impairments in fetal alcohol spectrum disorder. JADR: (In press).
- McKay BE, Turner RW (2005) Physiological and morphological development of the rat cerebellar Purkinje cell. J Physiol 567: 829-850.
- Altman J (1972) Postnatal development of the cerebellar cortex in the rat. II. Phases in the maturation of Purkinje cells and of the molecular layer. J Comp Neurol 145: 399-463.
- Altman J (1972) Postnatal development of the cerebellar cortex in the rat. 3. Maturation of the components of the granular layer. J Comp Neurol 145: 465-513.
- Altman J (1972) Postnatal development of the cerebellar cortex in the rat. I. The external germinal layer and the transitional molecular layer. J Comp Neurol 145: 353-397.
- Hamano K, Takeya T, Iwasaki N, Nakayama J, Ohto T, et al. (1998) A quantitative study of the progress of myelination in the rat central nervous system, using the immunohistochemical method for proteolipid protein. Brain Res Dev Brain Res 108: 287-293.
- D'Ercole AJ, Ye P, Calikoglu AS, Gutierrez-Ospina G (1996) The role of the insulin-like growth factors in the central nervous system. Mol Neurobiol 13: 227-255.
- de la Monte SM, Longato L, Tong M, Wands JR (2009) Insulin resistance and neurodegeneration: roles of obesity, type 2 diabetes mellitus and non-alcoholic steatohepatitis. Curr Opin Investig Drugs 10: 1049-1060.
- Zheng WH, Kar S, Doré S, Quirion R (2000) Insulin-like growth factor-1 (IGF-1): a neuroprotective trophic factor acting via the Akt kinase pathway. J Neural Transm Suppl : 261-272.
- Park M, Shen K (2012) WNTs in synapse formation and neuronal circuitry. EMBO J 31: 2697-2704.
- Ataman B, Ashley J, Gorczyca M, Ramachandran P, Fouquet W, et al. (2008) Rapid activity-dependent modifications in synaptic structure and function require bidirectional Wnt signaling. Neuron 57: 705-718.
- Krylova O, Herreros J, Cleverley KE, Ehler E, Henriquez JP, et al. (2002) WNT-3, expressed by motoneurons, regulates terminal arborization of neurotrophin-3-responsive spinal sensory neurons. Neuron 35: 1043-1056.
- Hall AC, Lucas FR, Salinas PC (2000) Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell 100: 525-535.
- Lebel C, Rasmussen C, Wyper K, Walker L, Andrew G, et al. (2008) Brain diffusion abnormalities in children with fetal alcohol spectrum disorder. Alcohol Clin Exp Res 32: 1732-1740.
- Sowell ER, Johnson A, Kan E, Lu LH, Van Horn JD, et al. (2008) Mapping white matter integrity and neurobehavioral correlates in children with fetal alcohol spectrum disorders. J Neurosci 28: 1313-1319.
- Watari H, Born DE, Gleason CA (2006) Effects of first trimester binge alcohol exposure on developing white matter in fetal sheep. Pediatr Res 59: 560-564.
- Archibald SL, Fennema-Notestine C, Gamst A, Riley EP, Mattson SN, et al. (2001) Brain dysmorphology in individuals with severe prenatal alcohol exposure. Dev Med Child Neurol 43: 148-154.
- Medina AE (2011) Fetal alcohol spectrum disorders and abnormal neuronal plasticity. Neuroscientist 17: 274-287.
- Medina AE, Krahe TE (2008) Neocortical plasticity deficits in fetal alcohol spectrum disorders: lessons from barrel and visual cortex. J Neurosci Res 86: 256-263.
- Abiola M, Favier M, Christodoulou-Vafeiadou E, Pichard AL, Martelly I, et al. (2009) Activation of Wnt/beta-catenin signaling increases insulin sensitivity through a reciprocal regulation of Wnt10b and SREBP-1c in skeletal muscle cells. PLoS One 4: e8509.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14904
- [From(publication date):
June-2013 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 10345
- PDF downloads : 4559