Research Article Open Access
Chitin Nanowhiskers Mediate Transformation of Escherichia coli by Exogenous Plasmid DNA
Alisa Mera1, Jun Araki2, Takashi Ohtsuki3, Makoto Shimosaka4 and Naoto Yoshida1*1Department of Biochemistry and Applied Biosciences, University of Miyazaki, 1-1 Gakuen Kibanadai-Nishi, Miyazaki 889-2192, Japan
2International Young Researchers Empowerment Center, Shinshu University, Tokida 3-15-1 Ueda, Nagano 386-8567, Japan
3Graduate School of Medicine and Engineering, University of Yamanashi, Kofu 400-8511, Japan
4Division of Applied Biology, Faculty of Textile Science and Technology, Shinshu University, Tokida 3-15-1, Ueda, Nagano 386-8567, Japan
- Corresponding Author:
- Naoto Yoshida, PhD
Department of Biochemistry and Applied Biosciences
University of Miyazaki, 1-1 Gakuen Kibanadai-Nishi
Miyazaki 889-2192, Japan
Tel: +81-985-58-7218
E-mail: a04109u@cc.miyazaki-u.ac.jp
Received date: September 03, 2011; Accepted date: September 26, 2011; Published date: September 28, 2011
Citation: Mera A, Araki J, Ohtsuki T, Shimosaka M, Yoshida N (2011) Chitin Nanowhiskers Mediate Transformation of Escherichia coli by Exogenous Plasmid DNA. J Biotechnol Biomaterial 1:114. doi:10.4172/2155-952X.1000114
Copyright: © 2011 Mera A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
A colloidal solution of chitin nanowhiskers containing pUC18 plasmid DNA and cells of Escherichia coli was placed on an agar hydrogel and stimulated by sliding friction applied between the agar hydrogel and polystyrene stir stick, leading to transformation of the bacteria to antibiotic resistance. The combination of chitin nanowhiskers and sliding friction was necessary for genetic transformation, indicating that the chitin nanowhiskers induced the Yoshida effect, resulting in the formation of E. coli cells penetration-intermediates. The pUC18 adsorbed onto the chitin nanowhiskers is presumably taken up through the penetration-intermediates, leading to transformation. The transformation efficiency changed when the number of recipient cells and amount of chitin nanowhiskers were varied. The maximum number of penetration-intermediates acquiring pUC18 DNA was obtained with concentrations of agar hydrogel, chitin nanowhiskers, and recipient E. coli cells of 2.5%, 5.0 ?g/ml, and 4.8 x 108 cells/ml, respectively. Optimal transformation efficiency with pUC18 was achieved with 2.1 x 1066 colony forming units/?g of pUC18. Induction of the Yoshida effect with chitin nanowhiskers represents a simpler alternative for introducing genes into bacteria.
Keywords
Chitin nanowhisker; Escherichia coli; Plasmid; Transformation; Yoshida effect
Introduction
Chitin is the most abundant natural amino polysaccharide, with more than 1011 tons of chitin produced annually [1]. There is currently great interest in chitin, not only as an underutilized resource, but also for its potential to serve as a valuable new multifunctional material. The current literature contains ample information regarding the purification [2,3], modification [4,5], degradation [6,7], and physical properties of chitin [8,9]. Because of their antibacterial, moisture retaining, and healing characteristics, chitin and chitosan (partially deacetylated chitin) are often utilized in water purification [10,11] or as additives in cosmetics [12,13], antibacterial agents [5,14], and pharmaceutical adjuvants [4,15,16]. However, many applications do not effectively utilize chitin, and a considerable amount goes to waste.
Heterogeneous acid hydrolysis of purified crab shell chitin produces fragmented microfibrils known as crystallites [17]. These partially acetylated crystallites are rod-like colloids produced by chain cleavage occurring at random locations along the microfibrils. Protonation of amino groups on the crystallites provides a positive surface charge and stabilizes the colloids due to repulsive forces between crystallites [18]. Li et al. [18] also reported that the degree of deacetylation, zeta potential, weight loss, and crystallinity of chitin nanowhiskers produced from crab shell chitin that has been deacetylated using a NaOH treatment can be controlled by manipulating the reaction time and conditions of acid hydrolysis.
When a colloidal solution consisting of nano-sized acicular material and bacterial cells is placed in a sliding friction field applied between a hydrogel and an interface forming material, the nano-sized acicular material and bacterial cells form a complex called a penetration-intermediate. This phenomenon is referred to as the Yoshida effect [19], and has been referenced by several microbiotechnology researchers [20,21]. Bacterial cells readily take up exogenous DNAs through penetrationintermediates by the Yoshida effect. Uptake of plasmid DNA adsorbed on chrysotile by the penetration-intermediate of Escherichia coli has been shown to alter its antibiotic resistance [22]. As a result of the discovery of the Yoshida effect, the penetration-intermediate has become a useful tool for introducing exogenous genes into bacteria [23], and for applications such as quantitative detection of asbestos species in the environment [24].
The occurrence of the Yoshida effect can be ascertained by assessing the transformation efficiency of the penetration-intermediate using plasmid DNA. The Yoshida effect has been confirmed using nanosized acicular materials including multi-walled carbon nanotubes, maghemite (γ-Fe2O3), α-sepiolite, and chrysotile with a diameter of 10–50 nm. Whichever nano-sized material is employed, a hydrogel, an interface-forming material, and an energy source for generating sliding friction are essential to produce the Yoshida effect.
The shape of chitin nanowhiskers prepared by Li et al. [18] and Revol and Marchessault [25] closely resembles that of the nano-sized acicular materials that induce the Yoshida effect. In this study, we evaluated whether chitin nanowhiskers are sufficient to produce the Yoshida effect by investigating the frequency of plasmid uptake in penetrationintermediate E. coli cells. We also determined the optimum conditions needed to form penetration-intermediates capable of acquiring plas- mid DNA in order to establish a genetic transformation method in E. coli dependent on chitin nanowhiskers and the Yoshida effect.
Materials and Methods
Preparation of a chitin nanowhisker suspension
An aqueous suspension of chitin nanowhiskers was prepared by acid hydrolysis of commercial a-chitin powder (Tokyo Chemical Industry Co., Ltd.) obtained from crab shells, according to a previously reported method [25]. Typically, 10 g of air-dried a-chitin powder was treated for 90 min with 100 ml of boiling 3 M HCl under reflux and with vigorous stirring. The sample was then washed with distilled water by repeated centrifugation (1600 x g, 5 min)-dilution cycles until the supernatant reached a pH of about 2. Above pH 2, the coarse aggregations from the hydrolysis residue began to disperse to yield a turbid supernatant containing colloidal particles of chitin nanowhiskers. Combined aliquots of the turbid supernatants from several centrifugation cycles were thoroughly dialyzed against distilled water until reaching a neutral pH, yielding a suspension with a solid content of 0.5–1%. The yield of nanowhiskers relative to the amount of starting a-chitin powder was around 70%.
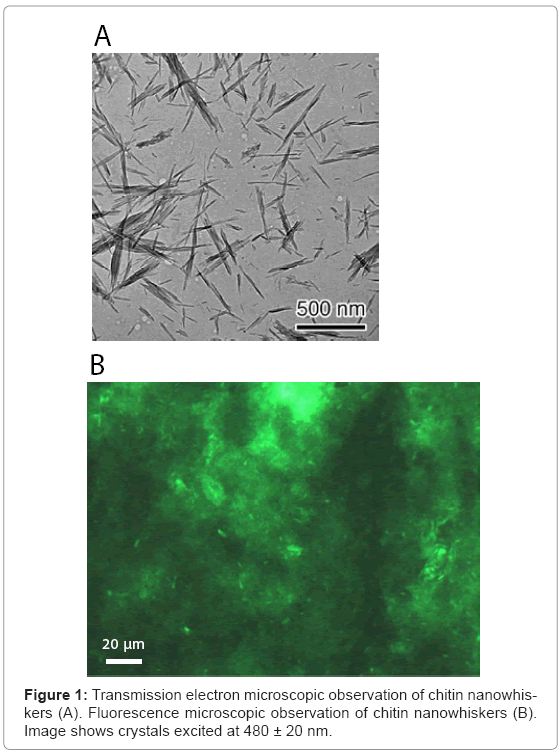
Transmission electron microscopy of chitin nanowhiskers
The morphology of the chitin nanowhiskers was observed using transmission electron microscopy (TEM). A drop of a dilute (< 0.1%) suspension of chitin nanowhiskers was deposited on a carbon-coated Formvar sheet spread on a copper grid (Okenshoji Co., Ltd., Tokyo, Japan). The dried sample was observed at 80 kV without staining and using a defocus contrast technique using JEOL JEM-2100 transmission electron microscope.
Fluorescent microscopic observation of chitin nanowhiskers
A chitin nanowhisker colloidal solution (2.8 mg/ml) was mixed with 0.5 μM synthetic DNA oligonucleotide (5’-CTA CCG CTT CGT GGA GCA GCC CGC CC-3’) tagged at the 5’ end with fluorescein isothiocyanate (FITC) (Sawady Co., Japan), and then observed directly using fluorescence microscopy (Axioskop2plus, Zeiss). For fluorescence studies, B-2A (BP 460–500 nm; DM 505 nm; BA 510–560 nm) filter blocks (Nikon) were used to visualize FITC. The experiments were documented using a Nikon digital camera.
Determining the amount of pUC18 adsorbed onto chitin nanowhiskers
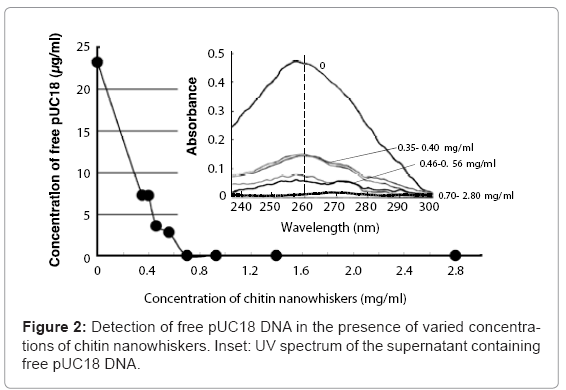
Colloidal solutions with varying concentrations of chitin nanowhiskers (0.35-1.40 mg/ml) were mixed with pUC18 DNA (at a final concentration of 25 μg/ml) and centrifuged at 15,000 x g for 10 min. The concentration of pUC18 DNA remaining in supernatant was determined using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, DE, USA) equipped with DNA measurement software. The wavelength spectrum of the supernatant was determined by wavelength scanning using the NanoDrop ND-1000 to confirm the presence of an absorbance peak at 260 nm. Wavelength scans were performed from 230 to 300 nm. The amount of pUC18 adsorbed onto the chitin nanowhiskers was calculated from the difference between the initial and the remaining amount of pUC18 in the supernatant.
Cultivation of recipient cells
Escherichia coli JM109 served as the recipient. Cells grown on Luria- Bertani (LB) plates [26] were inoculated into LB broth and cultured for 14–18 h at 30°C with aeration (provided by shaking at 150 rpm). The culture broth was directly subjected to the Yoshida effect when the cell density reached an OD550 of 3.0 (6.0 x 108 cells/ml), as determined by spectrophotometry.
Factors needed to induce the Yoshida effect
To investigate the occurrence of the Yoshida effect by using chitin nanowhiskers, Escherichia coli JM109, agar (Nacalai Tesque, Japan), polystyrene stir sticks (Sarstedt, Nünbrecht, Germany), and an automatic turning table equipped with a Gene Injector (Preci Co. Ltd., Japan) served as the plasmid recipient cells, the hydrogel, the interface forming material, and the energy source to produce a sliding friction field, respectively. pUC18 DNA was used as the donor plasmid. E. coli cells were transformed with pUC18 to express a phenotype of ampicillin resistance.
Standard protocol for inducing the Yoshida effect
Agar hydrogel plates were prepared by autoclaving a 1.5–3.5% agar (Nacalai Tesque, Japan) solution containing LB nutrient at 121°C for 15 min. Ampicillin was then added to a final concentration of 100 μg/ ml and the agar was allowed to solidify in Petri dishes (8.5 cm diameter). The surfaces of the agar hydrogel plates were rapidly dried in a clean room to remove all visible water condensation before applying sliding friction. A 50 μl volume of chitin nanowhisker colloidal solution (0.1–50 μg/ml) containing 1.0 ng of pUC18 DNA was placed on the agar hydrogel, followed by 50 μl of E. coli culture broth. Sliding friction was applied to the surface of the agar hydrogel without disrupting the agar for 60 seconds using a polystyrene stir stick by rotating the plate on an automatic tuning table set at 90 rpm. The vertical reaction force of the polystyrene stir stick was maintained at 40 g/cm2 using the Gene Injector [23].
Evaluation of the occurrence of the Yoshida effect
The frequency of penetration-intermediate formation is reflected by the number of colony forming units transformed to ampicillin resistance. After sliding friction was applied, the agar hydrogel plates were incubated at 37°C for 15–18 hr, after which the number of colonies demonstrating ampicillin resistance was determined. The transformation efficiency was expressed as the number of transformant colonies per microgram of pUC18 DNA.
Results
Transmission electron microscopy of chitin nanowhiskers
The colloidal chitin nanowhiskers obtained from crab chitin hydrolyzed with 3 M HCl was observed under a scanning electron microscope. As shown in Figure 1A, the presence of acicular and high crystallinity particles with an axial ratio of about 21 was confirmed. As the average size of a typical chitin nanowhisker was 7 x 7 x 150 nm, the number of chitin nanowhiskers was estimated to be approximately 9.55 x 1013 per mg.
Observation of chitin nanowhiskers using fluorescence microscopy
Chitin nanowhiskers mixed with DNA oligonucleotide tagged at the 5’ end with FITC were observed under a fluorescence microscope. Chitin nanowhiskers fluoresced green (Figure 1B). It should be noted that significant amount of chitin nanowhiskers aggregated themselves, but acicular crystalline material was also observed. These results indi cated that nucleic acid was effectively adsorbed onto the surface of the chitin nanowhiskers.
Amount of pUC18 DNA adsorbed onto chitin nanowhiskers
Mixing pUC18 DNA with chitin nanowhiskers resulted in adsorption of pUC18 onto the nanowhisker surfaces and a decrease in the amount of free pUC18. The pUC18 solution (23.5 μg/ml) solution without chitin nanowhiskers showed a large adsorption peak at 260 nm (Figure 2, inset). This peak decreased as the chitin nanowhisker concentration increased, and the DNA concentration in the supernatant was 3.55 and 2.78 μg/ml when the concentration of chitin nanowhisker was 0.46 and 0.56 mg/ml, respectively. No free pUC18 DNA was detected in the supernatant containing 0.70 mg/ml or more of chitin nanowhiskers (Figure 2). From these results, the maximum amount of pUC18 adsorbed onto the chitin nanowhiskers was calculated to be 35.7 μg/mg of chitin nanowhiskers.
Occurrence of the Yoshida effect induced by chitin nanowhiskers
When 50 μl of a mixture containing 30 μg/ml of chitin nanowhiskers, 200 ng/ml of donor pUC18, and 50 μl of E. coli culture broth was placed in a sliding friction field applied between an agar hydrogel (2.5%) and polystyrene stir stick, 1.5 x 104 colonies transformed to ampicillin resistance were obtained per microgram of pUC18 (Table 1). Under similar conditions but lacking either chitin nano whiskers or pUC18 DNA, no ampicillin resistant colonies appeared. These results indicated that the transformation of the E. coli cells to antibiotic resistance was not due to chemical mutation induced by the chitin nanowhiskers. Instead, intracellular uptake of exogenous pUC18 facilitated the transformation of the E. coli cells to antibiotic resistance. Similarly, an elimination of E. coli cells did not bring any transformed colonies, indicating that the E. coli transformants observed in the previous experiments were not environmental contaminants. As an additional control, a removal of sliding friction was found to produce no ampicillin resistant colonies. It is clearly shown that the combination of chitin nanowhiskers and sliding friction are essential for E. coli transformation with pUC18 DNA. The occurrence of genetic transformation of E.coli through the intracellular acquisition of pUC18 indicated that a penetration-intermediate was formed and that chitin nanowhiskers had induced the Yoshida effect in the E. coli cells. Next, we optimized the conditions to maximize the transformation efficiency with pUC18 in E. coli.
| Chitin nanowhisker (30 μg/ml) | pUC18 (200 ng/ml) | Recipient cell | Sliding friction | Transformation efficiency (cfu/μg) |
| + | + | + | + | 1.5E+04 |
| - | + | + | + | ND |
| + | - | + | + | ND |
| + | + | - | + | ND |
| + | + | + | - | ND |
cfu = colony forming units
ND = Not detected
Table 1: Combination of factors needed to introduce donor pUC18 DNA into recipient E. coli cells.
Effect of hydrogel hardness on transformation efficiency
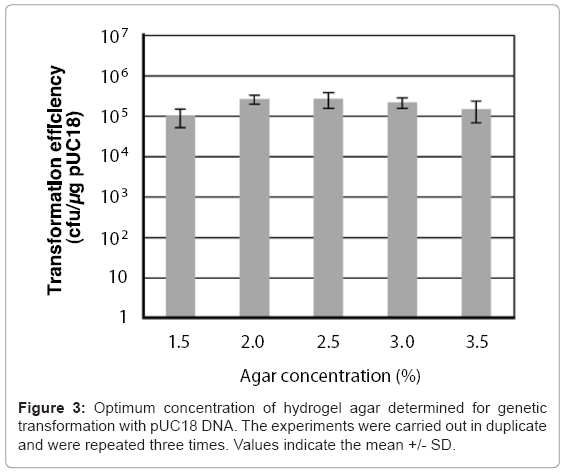
A 50 μl chitin nanowhisker colloidal solution containing pUC18 and 50 μl of E. coli culture broth was placed on variable concentration (1.5–3.5%) agar hydrogels, after which sliding friction was applied to the hydrogel surface. The concentrations of chitin nanowhiskers and E. coli cells were 30 μg/ml and 6.0 x 108 cells/ml, respectively. As shown in Figure 3, all concentrations of the agar hydrogel enabled the penetration- intermediate to acquire pUC18 with a transformation efficiency of more than 105 cells/μg of pUC18. The 2.5% agar provided the optimal hardness for transformation of the penetration-intermediate, producing a transformation efficiency of 2.6 x 105 cells/μg of pUC18.
Effect of chitin nanowhisker concentration on transformation efficiency
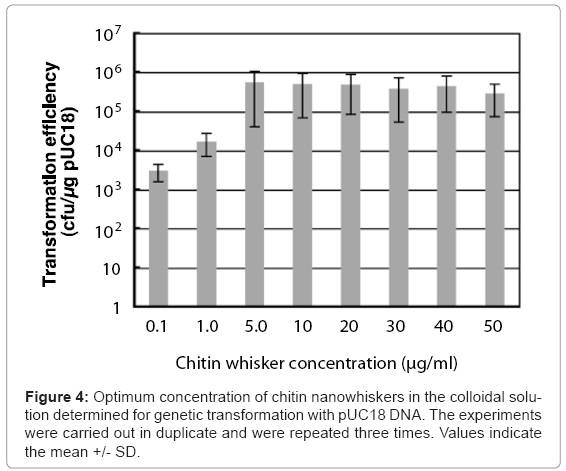
Colloidal solutions (50 μl) containing pUC18 and different concentrations of chitin nanowhiskers (0–150 μg/ml) were placed on agar hydrogels with 50 μl of E. coli culture broth, after which sliding friction was applied. The concentrations of agar and E. coli cells were 2.5% and 6.0 x 108 cells/ml, respectively. As shown in Figure 4, at chitin nanowhisker concentrations of 0.1 and 1.0 μg/ml, the transformation efficiency of the penetration-intermediate was relatively low, with 3 x 103 and 1.7 x 104 ampicillin-resistant colonies formed/μg of pUC18, respectively. On the other hand, at chitin nanowhisker concentrations between 5.0 and 50 μg/ml, the transformation efficiency rose to more than 105 ampicillin-resistant cells formed/μg of pUC18. The colloidal solution containing 5.0 μg/ml of chitin nanowhiskers was optimal for transformation of E. coli cells, resulting in a transformation efficiency of 5.0 x 105 ampicillin-resistant colonies formed/μg of pUC18.
Effect of recipient E. coli cell concentration on transformation efficiency
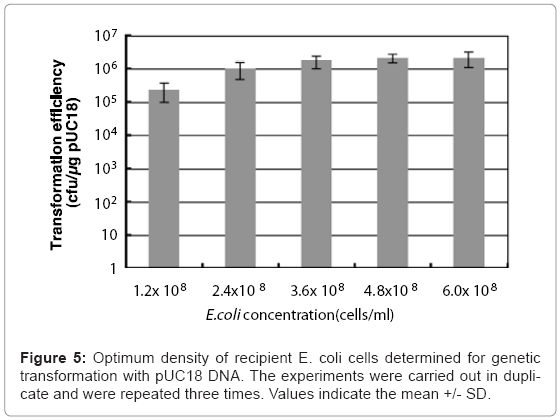
Colloidal solutions (50 μl) of chitin nanowhiskers (5.0 μg/ ml) containing pUC18 were placed on agar hydrogel with 50 μl of culture broth containing varying the concentration of E. coli cells from 1.2 x 108 to 6.0 x 108 cells/ml, after which sliding friction was applied. The optimal culture broth concentration of E. coli for transformation of the penetration- intermediate is 4.8 x 108 cells/ml, which represents 2.4 x 107 cells in 50 μl of culture broth. This concentration of E. coli cells produced a transformation efficiency of 1.2 x 106 ampicillin-resistant colonies formed/μg of pUC18. (Figure 5).
Discussion
When a colloidal solution consisting of nano-sized acicular material and bacterial cells is stimulated by sliding friction at the interface between the hydrogel and the interface-forming material, where the frictional coefficient rapidly increases, the nano-sized acicular material forms a chestnut–bur shaped complex. This complex grows larger and penetrates the bacterial cells due to the driving force derived from the sliding friction, a process known as the Yoshida effect [27]. Chrysotile and α-sepiolite are nano-sized acicular materials that have been used for transformation with plasmid DNA through the Yoshida effect. The ability of chrysotile and α-sepiolite to adsorb DNA was demonstrated by Yoshida and Ide [22]. We mixed chitin nanowhiskers with oligonucleotide tagged at the 5’ end with FITC and observed them under a fluorescence microscope. This experiment indicated that the chitin nanowhiskers have a high affinity for adsorbing nucleic acid that makes them suitable for transformation using the Yoshida effect (Figure 1B).
Xu et al. [28] reported that the positive charge of chitosan acts to condense the relatively large negative charge of the plasmid DNA, such that it can be incorporated into the chitosan nanoparticles. Incorporation of the plasmid into the chitosan nanoparticles does not affect the structural integrity of the plasmid, as demonstrated by gel electrophoresis. The chitin nanowhisker surface leaves amino groups due to acid hydrolysis-induced deacetylation. Conductimetric titration [29,30] indicated that acid hydrolysis-induced deacetylation resulted in 299 μmol of amino groups/g chitin nanowhiskers. The surface charge density of chitin nanowhiskers was demonstrated to be 0.45/nm2, which is close to the value of 0.5–0.6/nm2 previously reported by Li et al. [29]. The protonation of chitin nanowhisker surface amino groups imparts a positive surface charge that interacts with anionic polymers such as nucleic acids to form ionic complexes. Plasmid DNA adsorbs onto the surface of chitin nanowhiskers due to the negative charge of the DNA. The maximum amount of pUC18 DNA that will adsorb onto chrysotile whiskers at pH 7.0 is 1.1 μg/mg of chrysotile [22]. In contrast, the maximum amount of pUC18 DNA that adsorbs onto chitin nanowhiskers at pH 7.0 is estimated to be 35.7μg/mg of chitin nanowhiskers. The amount of nucleic acid that would adsorb onto chitin nanowhiskers is significantly greater than that adsorbs onto chrysotile whiskers. Our experiments reveal that chitin nanowhiskers form a chestnut–bur shaped complex that grows larger as a result of sliding friction and penetrates E. coli cells due to the driving force derived from the sliding friction. Plasmid DNA adsorbed onto the chitin nanowhiskers is then introduced into the bacterial cells through the perforations.
Several groups have reported genetic transduction using chitin, including Richardson et al. [31], Mao et al. [32], and Bozkir and Saka [33]. Bozkir and Saka [33] demonstrated that deacetylated chitin nanoparticles substituting for virus particles are safe for use as gene therapy delivery vehicles. Plasmid DNA was capsulated in 450–820 nm chitin nanoparticles prepared according to the solvent evaporation and complex coacervation methods. The authors confirmed that the chitin nanoparticles protect the encapsulated plasmid DNA from nuclease attack, and also reported that the efficiency of chitin nanoparticle-mediated transformation of E. coli is significantly higher than transformation with naked DNA. However, they did not report the efficiency of transforming E. coli with plasmid DNA.
The efficient introduction of DNA into bacteria is a phenomenon of great practical importance in genetic engineering and molecular biology. The introduction of exogenous DNA into E. coli was first demonstrated by Mandel and Higa [34], who observed that incubating a suspension of E. coli cells and bacteriaophage lDNA in a solution of CaCl2 at 0°C resulted in the subsequent appearance of infectious centers. They further showed that a heat pulse, in which the mixture of cells and DNA was briefly incubated at 42°C, chilled on ice, and then diluted into growth medium, improved the frequency of transfection. Since the first demonstration of Ca2+-dependent DNA transfer into E. coli by Cohen [35], Hanahan’s protocol has been rigorously developed to achieve maximum transformation efficiency, and is now one of the best chemical transformation methods available [36,37]. This method has been successfully applied to E. coli strain DH5α, yielding transformation efficiencies of 105–109 per microgram of plasmid DNA. We determined that E. coli JM109 can be transformed to ampicillin resistance via plasmid DNA and chitin nanowhiskers through the Yoshida effect, with a maximum transformation efficiency of 2.1 x 109 per microgram of pUC18. The transformation efficiency we achieved using chitin nanowhiskers was comparable to that obtained using conventional chemical methods.
A great deal of effort has gone into the development of transformation technology, resulting in a large body of knowledge and a diverse array of methodologies. Some methods are very simple but are not widely applicable. Other methods may be more widely applicable but are more complex or difficult. Our newly developed method is simple, reliable, and highly reproducible, with the additional benefit that plating and transformation occur simultaneously. Interest in using chitin nanowhiskers and sliding friction force for gene introduction rather than chemical transformation is certain to increase as more researchers become aware of the method’s low toxicity and technical simplicity.
Acknowledgements
The authors would like to thank Mr. Yuta Yamanaka of Shinshu University for technical contributions regarding conductometric titration of chitin nanowhisker surface amino groups. One of the authors (MS) is grateful to the Institute for Fermentation, Osaka for providing a Research Grant.
References
- Yu C, Bassler BL, Rosema, S (1993) Chemotaxis of the marine bacterium Vibrio furnissii to sugars: a potential mechanism for initiating the chitin catabolic cascade. J Biol Chem 268: 9405-9409.
- Charoenvuttitham P, Shi J, Gauri M (2006) Chitin extraction from black tiger shrimp (Penaeus monodon) waste using organic acids. Separ Sci Tech 41: 1135-1153.
- Xu Y, Gallert C, Winter J (2008) Chitin purification from shrimp wastes by microbial deproteination and decalcification. Appl Microbiol Biotechnol 79: 687-697.
- Jayakumar R, New N, Tokura S, Tamura H (2007) Sulfated chitin and chitosan as novel biomaterials. Int J Biol Macromol 40: 175-181.
- Jiang X, Cai K, Zhang J, Shen Y, Wang S, et al. (2011) Synthesis of a novel water-soluble chitosan derivative for flocculated decolorization. J Hazard Mater 185: 1482-1488.
- Imai T, Watanabe T, Yui T, Sugiyama J (2002) Directional degradation of betachitin by chitinase A1 revealed by a novel reducing end labelling technique. FEBS Lett 510: 201-205.
- Taghizadeh MT, Abdollahi R (2011) Sonolytic, sonocatalytic and sonophotocatalytic degradation of chitosan in the presence of TiO2 nanoparticles. Ultrason Sonochem 18: 149-157.
- Noh HK, Lee SW, Kim JM, Oh JE, Kim KH, et al. (2006) Electrospinning of chitinnanowhiskers: degradation behavior and cellular response to normal human keratinocytes and fibroblasts. Biomaterials 27: 3934-3944.
- Onsøyen E, Skaugrud O (1990) Metal recovery using chitosan. J Chem Technol Biotechnol 49: 395-404.
- Agerkvist I, Eriksson L, Enfors SO (1990) Selective flocculation with chitosan in Escherichia coli disintegrates: effects of pH and nuclease treatment. Enzyme Microb Technol 12: 584-590.
- Wang JP, Chen YZ, Yuan SJ, Sheng GP, Yu HQ (2009) Synthesis and characterization of a novel cationic chitosan-based flocculant with a high watersolubility for pulp mill wastewater treatment. Water Res 43: 5267-5275.
- Anchisi C, Meloni MC, Maccioni AM (2006) Chitosan beads loaded with essential oils in cosmetic formulations. J Cosmet Sci 57: 205-214.
- Gautier S, Xhauflaire-Uhoda E, Gonry P, Piérard GE (2008) Chitin-glucan, a natural cell scaffold for skin moisturization and rejuvenation. Int J Cosmet Sci 30: 459-469.
- Potara M, Jakab E, Damert A, Popescu O, Canpean V, et al. (2011) Synergistic antibacterial activity of chitosan-silver nanocomposites on Staphylococcus aureus. Nanotechnology 22: 135101.
- Baldrick P (2010) The safety of chitosan as a pharmaceutical excipient. Regul Toxicol Pharmacol 56: 290-299.
- Muzzarelli RA, Morganti P, Morganti G, Palombo P, Palombo M, et al. (2007) Chitin nanofibrils/chitosan glycolate composites as wound medicaments. Carbohydr Polym 70: 274-284.
- Desai K, Kit K, Li J, Zivanovic S (2008) Morphological and surface properties of electrospun chitosan nanofibers. Biomacromolecules 9: 1000-1006.
- Li J, Revol JF, Marchessault RH (1998) Effect of degree of deacetylation of chitin on the properties of chitin crystallites. J Appl Polymer Sci 65: 373-380.
- Yoshida N (2007) Discovery and application of the Yoshida effect: nano-sized acicular materials enable penetration of bacterial cells by sliding friction force. Recent Pat Biotechnol 1: 194-201.
- Aune TE, Aachmann FL (2010) Methodologies to increase the transformation efficiencies and the range of bacteria that can be transformed. Appl Microbiol Biotechnol 85: 1301-1313.
- Wilharm G, Lepka D, Faber F, Hofmann J, Kerrinnes T, et al. (2010) A simple and rapid method of bacterial transformation. J Microbiol Methods 80: 215-216.
- Yoshida N, Ide K (2008) Plasmid DNA is released from nanosized acicular material surface by low molecular weight oligonucleotides: exogenous plasmid acquisition mechanism for penetration intermediates based on the Yoshida effect. Appl Microbiol Biotechnol 80: 813-821.
- Yoshida N, Nakajima-Kambe T, Matsuki K, Shigeno T (2007) Novel plasmid transformation method mediated by chrysotile, sliding friction, and elastic body exposure. Anal Chem Insights 2: 9-15.
- Yoshida N, Takebe K (2006) Quantitative detection of asbestos fiber in gravelly sand using elastic body-exposure method. J Ind Microbiol Biotechnol 33: 827-833.
- Revol J-F, Marchessauld RH (1993) In vitro chiral nematic ordering of chitin crystallites. Int J Biol Macromol 15: 329-335.
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edition Cold Spring Harbor Laboratory, New York.
- Yoshida N, Saeki Y (2004) Chestnut bur-shaped aggregates of chrysotile particles enable inoculation of Escherichia coli cells with plasmid DNA. Appl Microbiol Biotechnol 65: 566-575.
- Xu X, Capito RM, Spector M (2007) Plasmid size influences chitosan nanoparticle mediated gene transfer to chondrocytes. J Biomed Mater Res A 84: 1038- 1048.
- Li J, Revol JF, Marchessault RH (1996) Rheological properties of aqueous suspensions of chitin crystallites. J Colloid Interface Sci 183: 365-373.
- Li J, Revol JF, Naranjo E, Marchessault RH (1996) Effect of electrostatic interaction on phase separation behaviour of chitin crystallite suspensions. Int J Biol Macromol 18: 177-187.
- Richardson SC, Kolbe HV, Duncan R (1999) Potential of low molecular mass chitosan as a DNA delivery system: biocompatibility, body distribution and ability to complex and protect DNA. Int J Pharm 178: 231-243.
- Mao HQ, Roy K, Troung-Le VL, Janes KA, Lin KY, et al. (2001) Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release 70: 399-421.
- Bozkir A, Saka OM (2004) Chitosan-DNA nanoparticles: effect on DNA integrity, bacterial transformation and transfection efficiency. J Drug Target 12: 281-288.
- Mandel M, Higa A (1970) Calcium-dependent bacteriophage DNA infection. J Mol Biol 53: 159-162.
- Cohen SN, Chang C, Hsu L (1972) Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A 69: 2110-2114.
- Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557-580.
- Inoue H, Nojima H, Okayama H (1990) High efficiency transformation of Escherichia coli with plasmids. Gene 96: 23-28.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 15199
- [From(publication date):
October-2011 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 10524
- PDF downloads : 4675