Research Article Open Access
Characterization of Clostridium acetobutylicum Protein Interaction Network from Butanol Fermentation
| Kumaran Sivagnanam1, Vijaya G.S. Raghavan1, Manesh Shah2, Robert L Hettich2, Nathan C Verberkmoes2 and Mark G Lefsrud1* | ||
| 1Department of Bioresource Engineering, Macdonald Campus, McGill University, Quebec, Canada | ||
| 2Oak Ridge National Laboratory, Chemical and Life Sciences Divisions, Oak Ridge, TN, USA | ||
| Corresponding Author : | Dr. Mark G Lefsrud Department of Bioresource Engineering Macdonald Campus, McGill University Quebec, Canada E-mail: mark.lefsrud@mcgill.ca |
|
| Received March 29, 2012; Accepted June 14, 2012; Published June 19, 2012 | ||
| Citation: Sivagnanam K, Raghavan VGS, Shah M, Hettich RL, Verberkmoes NC, et al. (2012) Characterization of Clostridium Acetobutylicum Protein Interaction Network from Butanol Fermentation. J Anal Bioanal Tech S3:002. doi: 10.4172/2155-9872.S3-002 | ||
| Copyright: © 2012 Sivagnanam K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Successful industrial production of butanol requires an in depth knowledge of Clostridium acetobutylicum. Development of computational approaches for protein interaction studies enabled us to generate a predicted protein interaction network of C. acetobutylicum ATCC 824. In this study, over 400 proteins from C. acetobutylicum were identified during butanol fermentation using shotgun proteomics and from only 217 proteins, we predicted 1947 interactions. This analysis showed that C. acetobutylicum proteins form a highly interconnected network and the main represented functions were found to be protein synthesis, carbohydrate metabolism and butanoate metabolism. We further used this interaction data to analyse the enrichment of gene ontology terms and pathways in C. acetobutylicum. Our analysis revealed that biological processes such as translation, cellular protein metabolic process, biosynthetic process, and pathways including glycolysis, butanoate metabolism were significantly enriched in C. acetobutylicum during butanol fermentation. This study provides a novel insight into the functional mechanisms of C. acetobutylicum at the network level complementing the current knowledge in relation to this industrially important organism.
| Keywords | |
| C. acetobutylicum; Protein-protein interaction; ABE fermentation; Butanol; Shotgun proteomics | |
| Introduction | |
| Clostridium acetobutylicum is a gram positive, spore forming, strictly anaerobic bacterium, capable of converting carbohydrates into acetone, butanol and ethanol in the ratio of 3:6:1 through a fermentation process [1]. ABE (Acetone-butanol-ethanol) fermentation suffers from several limitations such as butanol toxicity and low solvent productivity and could no longer compete with the chemical synthesis of butanol [2]. However, depletion of oil reserves and the quest for renewable fuel has renewed interest in using butanol as a fuel and revived the research interest in the economically viable butanol production through ABE fermentation process [3]. Several approaches such as traditional mutagenesis and recombinant DNA technology have been employed to amend the targeted metabolic pathways in order to improve the performance of solventogenic clostridia [4]. Genetic and metabolic engineering of solventogenic clostridia have been performed with the aim of generating strains that can be used for enhanced butanol tolerance and production [5]. Although significant progress have been made over the past years, improvements in the solventogenic clostridia are necessary to develop a more competitive industrial scale butanol production process. Recent developments in genetics, genomics and proteomics research fields have greatly increased our understanding of C. acetobutylicum. The complete genome sequence of C. acetobutylicum ATCC 824 which consists of 3,940,880bp chromosome and a 192,000bp megaplasmid pSOL1 was published and a total of 3740 and 178 ORFs were identified on the chromosome and megaplasmid, respectively [6]. Following the reporting of the complete genome sequence, genomewide transcriptome studies on C. acetobutylicum were performed [7- 10] and a few proteomic studies [11-15] were carried out. However, a detailed knowledge of the complex, little-known metabolic network of C. acetobutylicum is essential to make a breakthrough in the metabolic engineering of this industrially important organism [16]. Protein interaction networks are key models to link molecules to biological functions [17] and it is of great interest to systematically map proteinprotein interactions as most of the cellular functions are mediated by groups of physically associated proteins or complexes that work in a coherent manner [18]. So far, no protein interaction network studies have been performed on this bacterium which constituted the aim of this study. In this paper, we describe a single large network of predicted protein-protein interactions for C. acetobutylicum. | |
| Materials and Methods | |
| Strain and fermentation development | |
| C. acetobutylicum ATCC 824 was obtained from American Type Culture Collection (ATCC, Cedarlane Labs, Ontario, Canada) and was cultured using reinforced clostridial medium (RCM) [19] in an anaerobic chamber (Coy Laboratory Products Inc., Michigan, US) at 37°C for 20-24h. Shake flask fermentation of C. acetobutylicum was performed in 250ml anaerobic flask containing 100ml of media consisting of (g/L) yeast extract (5.0), ammonium acetate (2.0), sodium chloride (1.0), KH2PO4 (0.75), K2HPO4 (0.75), cysteine HCl.H2O (0.50), MgSO4 (0.2), MnSO4.H2O (0.01), FeSO4.7H2O (0.01) and glucose (30.0) [20]. Shake flask fermentation were also performed using xylose with the same media composition except glucose. Before inoculation, the medium was autoclaved at 121°C for 15min (Cysteine HCl.H2O was filter sterilized through 0.45μm filter and added to the medium) and cooled to 35°C in anaerobic chamber. The cell suspension was incubated at 37°C with shaking at 120rpm and the growth was monitored with OD600nm. Agitation is required for maintaining the solid-liquid suspension homogeneous to ensure good mass transfer in and out of the microbial cell and to improve cell growth in anaerobic fermentations [21,22]. Samples of 10ml were harvested from the start of the inoculation in the fermentation experiment until to the stationary phase for further proteomic analysis. All chemicals used in this study were supplied from Fisher (Fisher Scientific, Canada) and Sigma (Sigma-Aldrich, Canada), unless otherwise specified. | |
| Product analysis | |
| Fermentation products (ABE, acetic acid and butyric acid) were analyzed by gas chromatography (Agilent 6890 series, Agilent Technologies, Wilmington, DE, USA) equipped with a HPINNOWax fused Capillary column (30 m × 0.25 mm, 0.25 μm, Agilent Technologies Inc., Quebec, CA) and a FID detector. The injector and detector temperatures were set at 220°C and 230°C, respectively. Column temperature was held at 150°C for 10 min and then increased by 15°C min−1 to 180°C, which was maintained for an additional 20 min. Nitrogen gas was used as a carrier gas at a pressure of 150 kPa [23]. ABE productivity was calculated as ABE produced in gL-1 divided by the fermentation time and is expressed as gL-1h-1. | |
| Cell lysis & protein extraction | |
| The microbial cell pellets (~100mg wet mass) from fermentation broth were processed through single tube whole cell lysis and protein digestion. Briefly, the cell pellet was resuspended in 1000μl of 6M guanidine/10mM Dithiothreitol (DTT) with 50mM Tris/10mM CaCl2 at pH 7.6 by vortexing every 10min for the first hour and incubated at 37°C for 12hrs to lyse cells and extrude proteins. The guanidine concentration was diluted with six-fold 50mM Tris buffer/10mM CaCl2 and 5-10μg sequencing grade trypsin (Promega, WI, USA) was added and incubated at 37°C for 12hrs to digest proteins to peptides. A second aliquot of the same amount of sequencing grade trypsin was added and incubated at 37°C for another 6hrs to ensure the digestion process. 1M DTT was added to a final concentration of 20mM and incubated for another hour with gentle rocking at 37°C. The complex peptide solution was centrifuged at 10,000g for 10min to remove cellular debris and the supernatant was collected, and cleaned using Sep-Pak plus (Waters Limited, Ontario, Canada). Using a Savant SpeedVac (Thermo Electron Corporation, Waltham, MA), samples was concentrated to ~200μl. For each LC-MS/MS analysis below, ~1/4 of the total sample was used based on the protocol followed by Verberkmoes [24]. | |
| Mass spectrometry | |
| Samples were analyzed in technical duplicates through a 2D nano- LC MS/MS system with a split-phase column [25] (~3-5cm SCX and 3-5cm C18) (Polymicro technologies, AZ) on a LTQ (ThermoFisher Scientific, CA, USA) with 12hr runs [26,27]. The LTQ settings were as follows: all data-dependent MS/MS in LTQ (top five), two microscans for both full and MS/MS scans, centroid data for all scans and two microscans averaged for each spectrum, dynamic exclusion set at 1. | |
| Proteome informatics | |
| All MS/MS spectra were searched with the SEQUEST algorithm [28] against C. acetobutylicum Uniprot proteome databases [29] and filtered with DTASelect/Contrast [25] at the peptide level (Xcorrs of at least 1.8 [+1], 2.5 [+2], 3.5 [+3]). Only proteins identified with two fully tryptic peptides from a 12h run were considered for further biological study. An in-house script was used to extract protein identifications, peptides, spectra, and sequence coverage from DTA Select filtered output files and used in calculation of protein abundance determination. | |
| Results and Discussion | |
| Fermentation | |
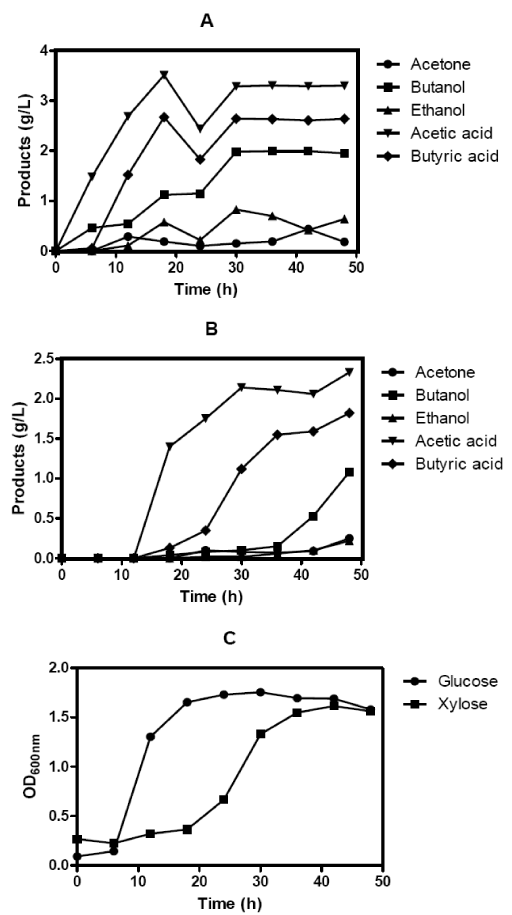
| ABE fermentation of C. acetobutylicum ATCC 824 using glucose and xylose substrate were examined. Growth profiles of the two substrates were documented by recording the Optical Density (OD) of biomass at 600nm and plotted against time. Glucose was found to be the preferred substrate for C.acetobutylicum with the total biomass concentration attaining the peak value of 1.76 in 30 h compared to the xylose with the total biomass concentration attaining the peak value of 1.61 in 42 h (Figure 1C). As reported by other authors [30], xylose is not preferred to glucose but did result in comparable cell density. After 48 h fermentation, the culture produced 2.77 g L-1 total ABE, resulting in an ABE productivity of 0.06 g L-1 h-1 for glucose substrate. Conversely, xylose utilized fermentation produced 1.55 gL-1 total ABE with productivity value of 0.03 g L-1h-1 (Figure 1A and Figure 1B). This demonstrated that C. acetobutylicum ATCC 824 utilized glucose as a preferred substrate compared to xylose for ABE fermentation. The results obtained here were in accordance with the literatures which reported that the efficiency of C. acetobutylicum in xylose utilization and solvent production are significantly lower when compared with glucose [30,31]. Based on the fermentation experiments, we expected that C. acetobutylicum proteins involved in the substrate utilization and solvent production pathways can be studied better in glucose utilized ABE fermentation than xylose. Therefore, the following C. acetobutylicum proteomic analysis was carried out from the samples collected at the late exponential phase of glucose utilized ABE fermentation. | |
| Shotgun proteomics approach | |
| Our results present the large scale investigation of the C. acetobutylicum proteome from a single time data point from the ABE fermentation process using glucose substrate by shotgun proteomics approach. This shotgun approach enabled us to detect proteins by matching peptide mass data to available genome sequence databases. All proteins in the non-redundant Uniprot proteome database [http:// www.uniprot.org] using keyword “C. acetobutylicum” that could match with the same set of peptides were included in the protein list. A total of 452 and 397 proteins were identified in this first and second mass spectrometry (MS) runs, respectively (Table 1 and Table 2). Recently, we have reported the proteomic analysis of C. acetobutylicum by shotgun proteomics using 22 h Mass Spectrometry (MS) run [14]. In this study, we have performed a 12 h MS run for the identification and analysis of protein-protein interaction as a proof of concept to study the protein interaction networks in C. acetobutylicum. | |
| The overall False Discovery Rate (FDR) was estimated by doubling the number of peptides found from the reverse database and dividing the result by the total number of identified peptides from both real and reverse databases using the formula: % fal = 2[nrev/(nrev + nreal)] × 100 where % fal is the estimated false discovery rate, nrev is the number of peptides identified from the reverse database and nreal is the number of peptides identified from the real database [32, 33]. The FDR was calculated as 0.22% and 0.25% for the first and second MS runs respectively. The relative abundances of the proteins identified during the MS analysis were estimated by calculating the Normalized Spectral Abundance Factors (NSAF). The NSAF for a protein is the number of spectral counts (SpC, the total number of MS/MS spectra) identifying a protein, divided by the protein’s length (L), divided by the sum of SpC/L for all proteins in the experiment [34,35]. The entire lists of proteins were sorted by averaged NSAF across the sample and its technical run. Based on the prediction of NSAF values, five most abundant proteins found to be present in C. acetobutylicum during the glucose utilized ABE fermentation process includes heat shock protein, glyceraldehyde-3-phosphate dehydrogenase, chaperonin, phosphocarrier protein, and acetyl-CoA acetyltransferase. | |
| Protein interaction network | |
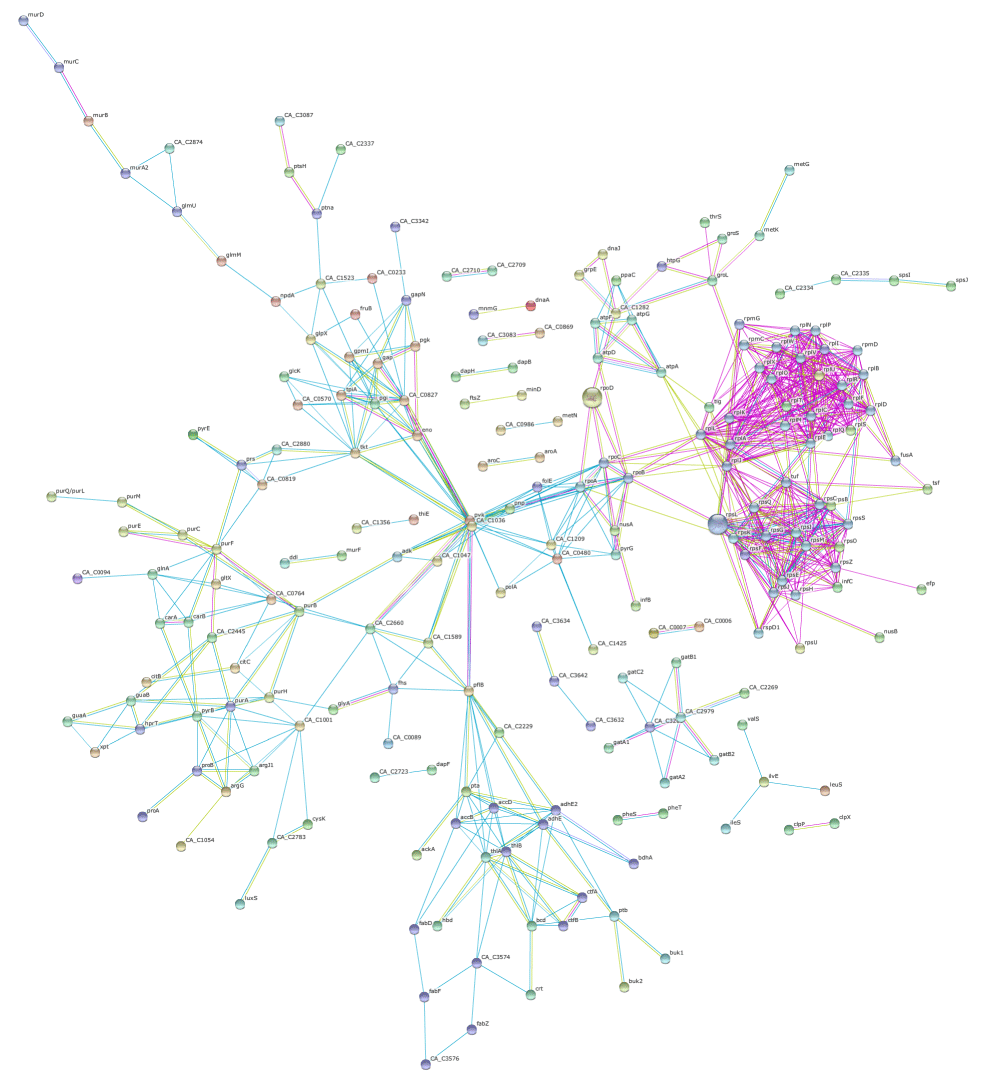
| The knowledge of C. acetobutylicum and its fermentation mechanism have been constantly increasing over the years. In parallel to the progress of C. acetobutylicum proteome studies, we used the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (version 9.0) tool (http://string.embl.de) [36] to assess the scope of proteinprotein interactions for the complete list of identified proteins from ABE fermentation using glucose. For data evaluation, the complete protein list was submitted as batch data and we applied “experiments”, “databases” and “text mining” as the prediction methods. To avoid spurious interactions in our large data set, we considered only the hits with the high stringency level with a confidence score of 0.7. The results are presented in Figure 2 (a scalable image is given in supplementary table) and a cluster of various interaction groups was immediately apparent from this interaction analysis. The most significant functions associated with this network were glycolysis, purine and pyrimidine metabolism, oxidative phosphorylation, butanoate metabolism, amino acid metabolism, transcription and translation. However, it is evident from the figure that a number of proteins were not interacted with any other protein. The major cluster of data set was comprised of ribosome proteins. Since ribosomes are involved in the process of protein synthesis, a range of transcription and translation associated proteins which include translation initiation factor, elongation factor, ribosomal proteins was found as a clustered group as well. The other highly connected interaction groups were composed of proteins involved in acid and solvent formation, and metabolisms such as sugars, amino acids and nucleotides. As can be inferred from Figure 2, proteins relating to carbohydrate metabolism, butanoate metabolism, amino acid and nucleotide metabolism as well as proteins involved in protein synthesis could be found to be tightly associated by the interaction analysis. | |
| Functional analysis of protein interaction network | |
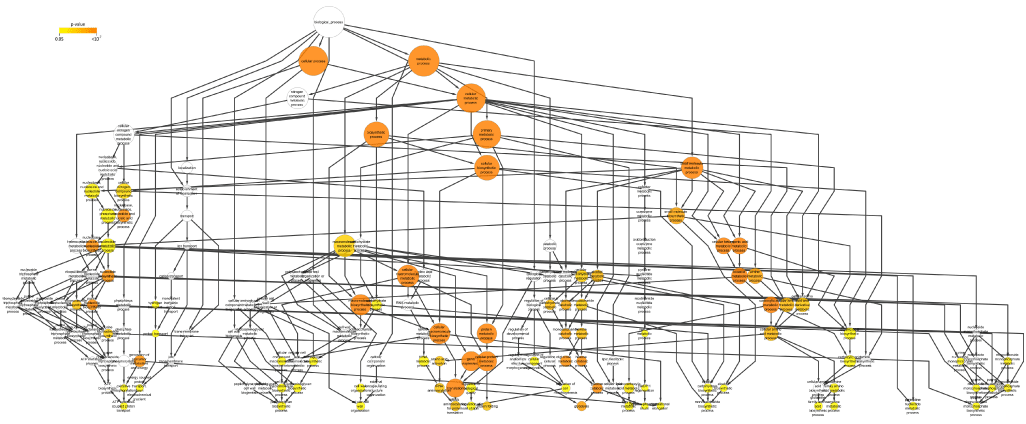
| The protein interaction network of the identified C. acetobutylicum proteins from STRING tool was loaded onto Cytoscape tool (version 2.8.2) [37] for functional protein annotation. The gene ontology (GO) annotations were obtained from the GO database (www.geneontology.org) and the BiNGO plugin (version 2.44) [38] of Cytoscape tool that assigns the functional annotations and scores them according to the enriched pathways. To gain insights into the functional roles of the identified C. acetobutylicum proteins and to highlight their functional mechanisms at the network level, the enriched GO terms were evaluated, specifically the biological process terms that describes the biological objectives to which the gene product contributes [39]. Functional analysis revealed that 89 biological process GO terms were over-represented (p<0.05) in the C. acetobutylicum protein interaction network (Supplementary Table). Figure 3 displays the distribution of functional categories in a hierarchical order which helps to cluster highly redundant/related terms typical of GO classification and detect related functional modules (a scalable image is given in supplementary table). | |
| The most highly represented GO biological processes (p<10-15) are related to translation (GO 6412), cellular protein metabolic processes (GO 44267), cellular biosynthetic process (GO 44249), cellular metabolic processes (GO 44237), and biosynthetic processes (GO 9058 and 34645) which are the basic processes common to all organisms. While the other highly enriched GO biological processes (p<10-5) are related to glycolysis (GO 6096), glucose metabolic process (GO 6006), hexose catabolic process (GO 19320) and carboxylic acid metabolic process (GO 19752) which are characteristic of C. acetobutylicum. These results correlate well with literatures which reported that C. acetobutyli-cum is capable of degrading a wide variety of carbohydrate substrates that are primarily catabolized through the glycolysis pathway in C. acetobutylicum [40,41]. Interestingly, the least enriched GO biological process terms include processes such as Pentose Phosphate Pathway (PPP) (GO 6098) and regulation of cell morphogenesis (GO 22604). This is in accordance with earlier studies which reported that fermentation of pentose sugars are repressed by hexoses in C. acetobutylicum [30,31,42]. | |
| In addition to BiNGO analysis, a similar method for the detection of enriched metabolic pathways in C. acetobutylicum was conducted using the DAVID program (http://david.abcc.ncifcrf.gov) [43]. The pathways that were significantly enriched in C. acetobutylicum during ABE fermentation were summarized in Table 2. The most enriched pathway (corrected p-value of 6.8e-14) was found to be the ribosome involved in basic processes such as translation and proteins synthesis which reiterates the results obtained from the BiNGO analysis. Other pathways that were found to be significantly enriched include butanoate metabolism (corrected p-value of 5.6e-02), pyruvate metabolism (corrected p-value of 5.9e-01) and glycolysis (corrected p-value of 5.9e-01). A detailed description of the C. acetobutylicum fermentation mechanism was well documented in literature which reported that C. acetobutylicum undergoes a biphasic fermentation process where acetate and butyrate acids were produced in the acidogenic phase and acetone, butanol and ethanol were produced in the solventogenic phase [5,44]. Therefore, the results obtained here suggest that the enriched pathways such as butanoate metabolism were critical for solvent formation and deregulation of these pathways leads to the loss of solvent production. | |
| Overall, we have established a single large network of protein interactions among C. acetobutylicum proteins identified from ABE fermentation. This study provides an insight of C. acetobutylicum fermentation characteristics and mechanism at the network level and will serves as a base for future investigations in relation to C. acetobutylicum protein interaction studies. | |
| Acknowledgements | |
| The ORNL part of this research was sponsored in part by U.S. Department of Energy under Contract DE-AC05-00OR22725 with Oak Ridge National Laboratory, managed and operated by UT-Battelle, LLC. We thank Dr. Stan Kubow and Dr. Kebba Sally, McGill University, for providing the gas chromatography to perform the fermentation product analysis. Special thanks to Manuel Ivan Villalobos Solis, McGill University for helping with the Cytoscape tool. | |
References
- Jones D, Woods DR (1986) Acetone-butanol fermentation revisited. Microbiol Rev 50: 484-524.
- Green E, Bennett GN (1996) Inactivation of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. Appl biochem biotechnol 57: 213-221.
- Gapes JR (2000) The economics of acetone-butanol fermentation: theoretical and market considerations. J Mol Microbiol Biotechnol 2: 27-32.
- Ezeji TC, Qureshi N, Blaschek HP (2007) Bioproduction of butanol from biomass: from genes to bioreactors. Curr Opin Biotechnol 18: 220-227.
- Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, et al. (2008) Fermentative butanol production by clostridia. Biotechnol Bioeng 101: 209-225.
- Nolling J, Breton G, Omelchenko M, Makarova K, Zeng Q, et al. (2001) Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol 183: 4823-4838.
- Tomas CA, Alsaker KV, Bonarius HP, Hendriksen WT, Yang H, et al. (2003) DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J Bacteriol 185: 4539-4547.
- Tummala SB, Junne SG, Paredes CJ, Papoutsakis ET (2003) Transcriptional analysis of product-concentration driven changes in cellular programs of recombinant Clostridium acetobutylicumstrains. Biotechnol Bioeng 84: 842-854.
- Alsaker KV, Spitzer TR, Papoutsakis ET (2004) Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell's response to butanol stress. J Bacteriol 186: 1959-1971.
- Paredes CJ, Rigoutsos I, Papoutsakis ET (2004) Transcriptional organization of the Clostridium acetobutylicum genome. Nucleic Acids Res 32: 1973-1981.
- Schaffer S, Isci N, Zickner B, Dürre P (2002) Changes in protein synthesis and identification of proteins specifically induced during solventogenesis in Clostridium acetobutylicum. Electrophoresis 23: 110-121.
- Sullivan L, Bennett GN (2006) Proteome analysis and comparison of Clostridium acetobutylicum ATCC 824 and Spo0A strain variants. J Ind Microbiol Biotechnol 33: 298-308.
- Mao S, Luo Y, Zhang T, Li J, Bao G, et al. (2010) Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield. J Proteome Res 9: 3046-3061.
- Sivagnanam K, Raghavan VG, Shah M, Hettich RL, Verberkmoes NC, et al. (2011) Comparative shotgun proteomic analysis of Clostridium acetobutylicum from butanol fermentation using glucose and xylose. Proteome Sci 9: 66.
- Sivagnanam K, Raghavan VGS, Shah M, Hettich RL, Verberkmoes NC, et al. (2012) Shotgun proteomic monitoring of Clostridium acetobutylicum during stationary phase of butanol fermentation using xylose and comparison with the exponential phase. J Ind Microbiol Biotechnol 39: 949-955.
- Zheng YN, Li LZ, Xian M, Ma YJ, Yang JM, et al. (2009) Problems with the microbial production of butanol. J Ind Microbiol Biotechnol 36: 1127-1138.
- Pawson T, Nash P (2003) Assembly of cell regulatory systems through protein interaction domains. Science's STKE 300: 445-452.
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, et al. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A 98: 4569-4574.
- Hibsch A, Grinsted E (1954) Methods for the growth and enumeration of anaerobic spore-formers from cheese, with observations on the effect of nisin. J Dairy Res 21: 101-110.
- Qureshi N, Meagher MM, Huang J, Hutkins RW (2001) Acetone butanol ethanol (ABE) recovery by pervaporation using silicalite–silicone composite membrane from fed-batch reactor of Clostridium acetobutylicum. J Memb Sci187: 93-102.
- Yerushalmi L, Volesky B (1985) Importance of agitation in acetone-butanol fermentation. Biotechnol Bioeng 27: 1297-1305.
- Cai X, Bennett GN (2011) Improving the Clostridium acetobutylicum butanol fermentation by engineering the strain for co-production of riboflavin. J Ind Microbiol Biotechnol 38: 1013-1025.
- Areesirisuk A, Laopaiboon L, Khongsay N, Laopaiboon P (2010) Improvement of gas chromatographic analysis for organic acids and solvents in acetone-butanol-ethanol fermentation from sweet sorghum juice. African Journal of Biotechnology 9: 6422-6429.
- Verberkmoes N, Russell A, Shah M, Godzik A, Rosenquist M, et al. (2008) Shotgun metaproteomics of the human distal gut microbiota. ISME J 3: 179-189.
- Tabb DL, McDonald WH, Yates JR 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21-26.
- Lo I, Denef VJ, VerBerkmoes NC, Shah MB, Goltsman D, et al. (2007) Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446: 537-541.
- Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, et al. (2005) Community proteomics of a natural microbial biofilm. Science 308: 1915-1920.
- Eng J, McCormack A, Yates JR 3rd (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976-989.
- Apweiler R, Bairoch A, Wu C, Barker W, Boeckmann B, et al. (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32: D115-D119.
- Ounine K, Petitdemange H, Raval G, Gay R (1985) Regulation and butanol inhibition of D-xylose and D-glucose uptake in Clostridium acetobutylicum. Appl environ microbiol 49: 874-878.
- Fond O, Engasser JM, Matta-El-Amouri G, Petitdemange H (1986) The acetone butanol fermentation on glucose and xylose. I. Regulation and kinetics in batch cultures. Biotechnol bioeng 28: 160-166.
- Mosley AL, Florens L, Wen Z, Washburn MP (2009) A label free quantitative proteomic analysis of the Saccharomyces cerevisiae nucleus. J proteomics 72: 110-120.
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC- MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2: 43-50.
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, et al. (2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40: 303-311.
- Zybailov BL, Florens L, Washburn MP (2007) Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol BioSyst 3: 354-360.
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, et al. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561-D568.
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366-2382.
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess over representation of gene ontology categories in biological networks. Bioinformatics 21: 3448-3449.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25-29.
- Steinmetz M (1993) Carbohydrate catabolism: pathways, enzymes, genetic regulation, and evolution. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics American Society for Microbiology, Washington, DC.
- Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, et al. (2002) Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Scis U S A 99: 996-1001.
- Fond O, Engasser JM, Matta-El-Amouri G, Petitdemange H (1986) The acetone butanol fermentation on glucose and xylose. II. Regulation and kinetics in fed-batch cultures. Biotechnol bioeng 28: 167-175.
- Huang da W, Sherman BT, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44-57.
- Dürre P (2005) Formation of solvents in clostridia. CRC Press, Boca Raton, FL, USA.
Tables and Figures at a glance
| Table 1 | Table 2 |
 |
 |
 |
||
| Figure 1 | Figure 2 | Figure 3 |
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14859
- [From(publication date):
August-2013 - Jul 08, 2025] - Breakdown by view type
- HTML page views : 10131
- PDF downloads : 4728
Peer Reviewed Journals
Make the best use of Scientific Research and information from our 700 + peer reviewed, Open Access Journals
