Research Article Open Access
Changes in the Matrix Markedly Enhance the Resolution and Accurate Identification of Human Pathogens by MALDI-TOF MS
Renata A. Culak, Min Fang, Shurene Bishop Simon, Itaru Dekio, Lakshani K. Rajakaruna and Haroun N. Shah*
Department for Bioanalysis and Horizon Technologies, Health Protection Agency Microbiology Services, London NW9 5EQ
- *Corresponding Author:
- Haroun N. Shah
Head, Molecular Identification Services Unit
Department for Bioanalysis and Horizon Technologies
Health Protection Agency Microbiology Services, Colindale
61 Colindale Avenue, London NW9 5EQ
Tel: 02083276749
E-mail: Haroun.shah@hpa.org.uk
Received date: March 29, 2012; Accepted date: May 11, 2012; Published date: May 16, 2012
Citation: Culak RA, Fang M, Simon SB, Dekio I, Rajakaruna LK, et al. (2012) Changes in the Matrix Markedly Enhance the Resolution and Accurate Identification of Human Pathogens by MALDI-TOF MS. J Anal Bioanal Tech S2:002. doi: 10.4172/2155-9872.S2-002
Copyright: © 2012 Culak RA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
The mass spectral profiles of three major/emerging Gram positive pathogens belonging to the genera Clostridium, Listeria and Propionibacterium were investigated using four MALDI-TOF (Matrix-Assisted Laser Desorption/ Ionization- Time of Flight) Mass Spectrometers from Waters, Shimadzu, Bruker and Ciphergen Biosystems. These instruments have been extensively used for developing a diagnostic platform for high throughput, low cost, near sample-free preparation for microbial identification. The results demonstrate the marked effect of spectral quality obtained by altering the matrix and the added value of simple preparative extraction procedures. While microbial spectral data are generally collected in the mass range 2 to 20 kDa for diagnostic signatures, most of the significant mass ions are <10 kDa. The change from the traditional use of CMBT (5-chloro-2-mercaptobenzothiazole) to DHB (2-(2, 5-DiHydroxyBenzoic acid) enabled successful identification of C. difficile whereas the substitution of CHCA (alpha-cyano-4-hydroxycinnamic acid) for DHB (2,5-dihydroxybenzoic acid) improved the identification of Listeria species. By contrast, the use of SELDI-TOF-MS (Surface Enhanced Laser Desorption/Ionization- Time of Flight- Mass Spectrometry), which employs affinity pre-capture of specific classes of molecules and analysed using sinapinic acid, showed a fivefold extension of the mass range of analytes, extending the potential for biomarker discovery and higher resolution strain typing of P. acnes.
Abbreviations
CBA: Colombia Blood Agar; FAA: Fastidious Anaerobe Agar; FAB: Fastidious Anaerobe Broth; NA: Nutrient Agar; BN: Blood Nutrient Agar; TFA: Trifluoroacetic acid; MALDI-TOF MS: Matrix Assisted Laser Desorption/Ionisation Time of Flight Mass Spectrometry; SELDI-TOF MS; Surface Enhanced Laser Desorption/ Ionisation Time of Flight Mass Spectrometry; CHCA: (alpha-cyano- 4-hydroxycinnamic acid); SPA: Sinapinic Acid; CMBT: 5-Chloro-2- MercaptoBenzoThiazole; DHB: (2,5-dihydroxybenzoic acid)
Introduction
A decade of intensive research based both on proteins and nucleic acids have seen Matrix Assisted Laser Desorption/Ionisation Time of Flight Mass Spectrometry (MALDI-TOF MS) gradually developed from an analytical tool to a diagnostic platform for identification of human pathogens [1-4]. Recent reports from diagnostic laboratories have shown that even among very fastidious groups of microorganisms, approximately 70% of clinical isolates can be confidently resolved to the species level using high throughput MALDI-TOF MS [5]. Failure to improve successful identification of the remaining taxa is often attributed to the lack of fully comprehensive microbial mass spectral databases for comparative analysis. However, many species are genetically so highly related that the input of additional spectra is unlikely to resolve such problems. Currently, mass ions from MALDITOF MS spectra are restricted to those less than 20 kDa, thus one possibility might be to extend the mass range for species that cannot be resolved by this imposed restriction. Several parameters may be altered to achieve this, the simplest perhaps being the introduction of different matrices.
The first reports of the use of MALDI-TOF MS for microbial identification demonstrated that alpha-cyano-4-hydroxycinnamic acid (CHCA) and 5-chloro-2-mercaptobenzothiazole (CMBT) were universally applicable for microorganisms [6,7]. Higher mass densities were achieved with CMBT and CHCA for Gram positive and Gram negative species respectively. Using this approach success was achieved for many poorly circumscribed anaerobic taxa [8] and the first comprehensive database of MS profiles was later reported [1]. However, there were exceptions to this binary approach, an important example being its inability to delineate isolates of the Gram positive human pathogen, Clostridium difficile. While this method successfully identified another major Gram positive nosocomial pathogen Staphylococcus aureus [9], C. difficile yielded such inconsistent and variable mass spectra that only 10% of the isolates were successfully identified [10]. This prompted indepth studies of the microbiological parameters that affect the mass spectral properties of this species with particular emphasis on the influence of the matrix. The present study reports the results of this work and demonstrates the marked effect upon the quality of a mass spectrum for the successful identification of this important human pathogen.
Another group included in the present study is the Gram positive genus Listeria.The genus comprises the species Listeria monocytogenes (type species), L. grayi, L. innocua, L. ivanovii, L. marthii, L. rocourtiae, L. seeligeri, and L. welshimeri. L. monocytogenes is pathogenic to man and a wide range of animals while another species, L. ivanovii primarily affects ruminants [11,12]. The remaining species are regarded as normal commensals. Infection by either pathogenic species may cause listeriosis, an infection previously considered to be very rare. Currently, listeriosis is associated with a mortality rate of between 20-40% and corroborates with a significant increase in hospitalisation rates from food-borne pathogens, with over 90% of cases being reported in Europe and the United States [13,14]. Because of the significant increase in the rates of infection now associated with L. monocytogenes, its unambiguous identification from other members of the genus is vital, therefore the effects of matrix and the use of preliminary extraction procedure to improve mass spectral resolution were investigated.
The third candidate species included in this study, Propionibacterium acnes, is a Gram positive, non-spore forming, facultatively anaerobic rod. P. acnes has long been considered one of the most dominant commensals of the human skin [15,16], however, there is a marked increase in P. acnes cases identified from a variety of opportunistic infections, such as acne vulgaris [17] (common acne), prosthetic hip joint infection [18], and prostate cancer [19]. P. acnes colonises an immensely varied range of ecological niches on the human skin. Consequently, it has been anticipated that its intra-species diversity may be high and numerous methods have been employed to ascertain whether there is any clonal selection of certain biotypes with infection. Three distinct types, type I, II, and III are currently recognised using molecular typing methods such as Multilocus Sequence Typing (MLST) based upon several housekeeping genes [20-22]. Type I is considered as the acne-specific subtype [20,23], while type II and III are considered to be as infectious in post-operative hip joint [20,21] Recently, it was shown that both type I and II share similar cell morphologies of short rods, while type III is filamentous [22,24]. Because P. acnes exhibits such a very complex population structure based on genome analysis, more in-depth analysis of its proteome may help delineate pathogenic from benign strains. For such an inherently diverse species, the need to increase its diagnostic mass range beyond 20 kDa is essential and has been explored here using affinity capture chip prior to MALDI-TOF MS (SELDI-TOF MS).
Materials and Methods
Cultures and maintenance
Culture Media: All media were supplied by Media Department, (Health Protection Agency Microbiology Services) HPA MS, Colindale.
a) C. difficile: Bacterial cultures (150 isolates) were maintained on both Colombia Blood Agar (CBA) and Fastidious Anaerobic Agar (FAA) for 24 h and 48 h at 37°C in anaerobic conditions. In addition, bacterial cultures were grown on Fastidious Anaerobic Broth (FAB) for 48 h at 37°C and re-subcultured on to FAA plates and incubated for 24 h at 37°C prior to MALDI-TOF MS analysis.
b) L. monocytogenes:The following 13 isolates were plated out (from beads stored at -20°C) onto Blood Nutrient (BN) Agar: L. monocytogenes NCTC 10357, L. monocytogenes F2365, L. monocytogenes NCTC 10888, L. monocytogenes NCTC 5348, L. monocytogenes 04883, L. monocytogenes NCTC 05124, L. grayi subsp. grayi NCTC 10815, L. grayi subsp. murrayi NCTC 10813, L. innocua NCTC 11288, L. ivanovii subsp. ivanovii NCTC 11846, L. ivanovii subsp. londoniensis NCTC 12701, L. seeligeri NCTC 11856, L. welshimeri NCTC 11857 and incubated overnight under aerobic conditions at 37°C. Purity was verified by visual inspection before use.
c) P. acnes: Forty bacterial cultures were maintained on CBA plates at 37°C under aerobic and anaerobic conditions (atmosphere containing 85% N2, 10% H2, 5% CO2) for 5 days. The source of these isolates have been reported previously [22].
Protein profiling of intact cells
MALDI Biotyper : A direct colony method for the preparation of bacteria was adopted in the study that involved the application of a single colony from growth plates directly onto a steel target plate as a thin film. Subsequently 1 μl of matrix solution (saturated solution of CHCA in 50% acetonitrile and 2.5% TFA) was applied to the colony and air dried. Mass spectra were acquired on a Bruker Biotyper system (Bruker Daltonics, Bremen, Germany). Positive charged ions were generated with a 337nm nitrogen laser, 240 spectra per sample were collected for each strain in a mass range of 2-20 kDa. Resulting spectra were analysed using MALDI Biotyper software (version 2.0, Bruker Daltonics, Bremen, Germany). Criteria for accurate identification used in the study were proposed by Bruker Daltonics: a score ≥ 2.000 indicated species level identification, a score of 1.700-1.999 indicated genus level identification, and a score < 1.700 was interpreted as no identification.
Reflectron M@LDI-TOF Mass Spectrometry: A small portion of a colony was removed from CBA plates and applied as a thin film to a stainless steel target plate using a 1 μl loop. 1 μl of matrix solution (saturated with CMBT in a solution containing acetonitrile, water and methanol in a ratio of 1:1:1 (v/v), 0.01M 18-crown-6 ether and 0.1% formic acid) was added to each spot. Mass profiles were generated using a Reflectron MALDI-TOF MS (Micromass, UK) equipped with a 337 nm nitrogen laser. Analysis was performed in positive-ion mode using a reflecting analyser with the voltage set at 2000 V.
Micrococcus lylae (NCTC 13377), EMRSA (NCTC 13134), MRSA (NCTC 11940), S. aureus (NCTC 7727), S. epidermidis (NCTC 11047) and S. epidermidis (NCTC 11407) were included as ‘blind’ controls. Mass spectra were acquired in batches of 12 replicates from each strain in a mass range of 0.5-10 kDa. Data processing was performed using MicrobeLynx™ software (Micromass, UK) and representative spectra of each test strain were compared to those already included in the software as described previously [1].
MALDI-TOF MS Axima™ – AnagnosTec- SuperSpectra™: Using a MALDI-TOF MS Axima™ (Shimadzu-Biotech Corp., Kyoto, Japan) and the identification system SARAMIS (Spectral Archive And Microbial Identification System, AnagnosTec GmbH, Potzdam, Germany), 150 isolates of C. difficile, grown under varying conditions and in different laboratories, were analysed. Mass fingerprints were obtained with detection in the linear, positive mode at a laser frequency of 50 Hz and within a mass range of 2 to 20 kDa. The acceleration voltage was 20 kV, and the extraction delay time was 200 ns. A minimum of 10 laser shots per sample was used to generate each ion spectrum. The wells of a FlexiMass MALDI target plates were smeared with cells using a 1 μl sterile loop and matrix solution (0.5 μl of 20 mg 2,5-dihydroxy benzoic acid dissolved in a 1 ml water-ethanol-acetonitrile [1:1:1] mix) was added to each sample and allowed to dry at room temperature. The Shimadzu Launchpad software program and the SARAMIS database (AnagnosTec GmbH) were used for automatic measurement and identification. Score values were determined by comparison against the ‘SuperSpectra’ for confident identification, from family to genus and species levels. A score ≥70% was considered a high confidence value for identification to the species level.
Protein profiling of cell extracts using SELDI-TOF Mass Spectrometry: SELDI-TOF mass spectrometry is unique in that it uses chromatographic surfaces to retain proteins based on their physiochemical characteristics. Proteins that bound and retained on the surfaces are analysed by mass spectrometry. However, whereas the three above methods utilised intact cells taken directly from a culture plate, analysis using SELDI-TOF-MS requires preparation of a cell lysate. The method described here was optimised for Propionibacterium acnes. Proteins were extracted using a standard buffer containing 8 M urea, 2 % CHAPS and 40 mM Trizma base. Each strain was grown for 3-5 days on three CBA plates and the cells harvested into 500 μl of the standard lysis buffer containing 300 mg of glass beads. The samples were then placed in a FastPrep device (MP Biologicals) for a total of 60 seconds. Samples were centrifuged at 21, 000 g for 30 min, supernatants collected and protein concentrations estimated using the standard Bradford assay.
The spots were pre-activated for 15 min with 5 μl of 20 mM Tris/ HCl pH 7.4 containing 0.1 % Triton X-100. The buffer was decanted and 6 μg of total protein in pre-activation buffer was applied onto each spot. The chip was left in a humidity chamber for 1 h. After the incubation, the unbound sample and the buffers were removed and the spots were washed three times with 20 mM Tris/HCl pH 7.4 containing 0.1 % Triton X-100. The chip was dried and 0.5 μl of sinapinic acid was applied twice onto each spot. The ProteinChip® arrays were analysed in a mass spectrometer (Ciphergen BioSystems, Model, PBS II) according to an automated data collection protocol [25]. The spectra were generated at laser intensity 220, high mass 50 kDa, detector sensitivity 10 and focus mass 25 kDa. The instrument was operated in positive ion mode and a nitrogen laser emitting at 337 nm was used.
Results
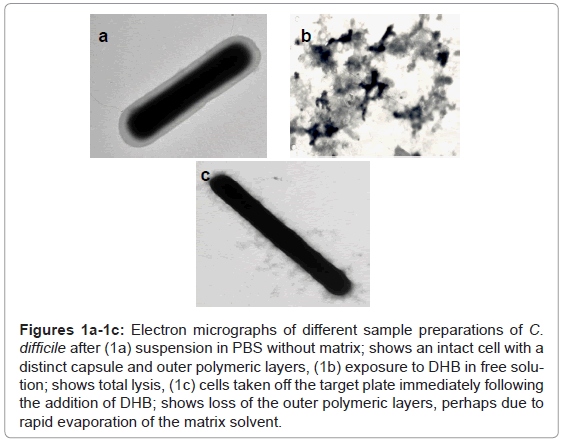
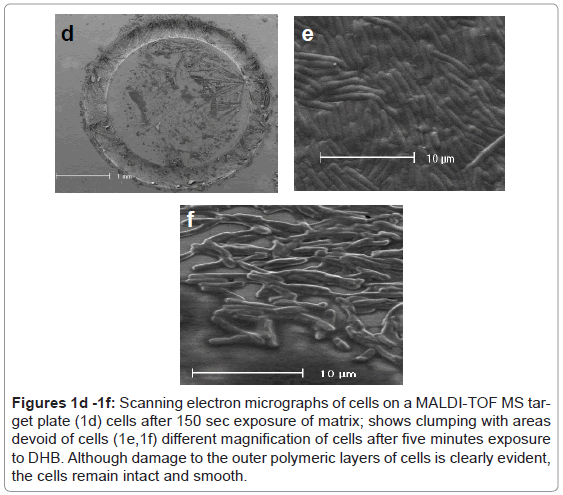
The substitution of DHB for CMBT as a matrix for the Gram positive species produced a marked effect upon the quality of the mass spectra obtained for C. difficile. Higher mass ion density was consistently achieved and this was not affected by the growth conditions used to maintain cells. Thus, cultures obtained from very different media, such as CBA, NA, FAA or FAB, yielded similar mass spectral profiles, thus the need to rigorously standardise this part of the protocol, as reported previously, was no longer essential [1]. Electron microscopy revealed that cells exposed to DHB in free solution immediately lysed compared those held in PBS (Figure 1a and Figure 1b). However, the integrity of the cell was partially retained if the cells were first smeared on the stainless target plate prior to addition of DHB (Figure 1c). Scanning electron micrographs showed the removal of the capsular polymers which perhaps facilitates ionisation of the abundant ribosomal proteins that are now known to comprise the key features of a bacterial mass spectrum (Figure 1d-1f).
Figures 1a-1c: Electron micrographs of different sample preparations of C. difficile after (1a) suspension in PBS without matrix; shows an intact cell with a distinct capsule and outer polymeric layers, (1b) exposure to DHB in free solution; shows total lysis, (1c) cells taken off the target plate immediately following the addition of DHB; shows loss of the outer polymeric layers, perhaps due to rapid evaporation of the matrix solvent.
Figures 1d -1f: Scanning electron micrographs of cells on a MALDI-TOF MS target plate (1d) cells after 150 sec exposure of matrix; shows clumping with areas devoid of cells (1e,1f) different magnification of cells after five minutes exposure to DHB. Although damage to the outer polymeric layers of cells is clearly evident, the cells remain intact and smooth.
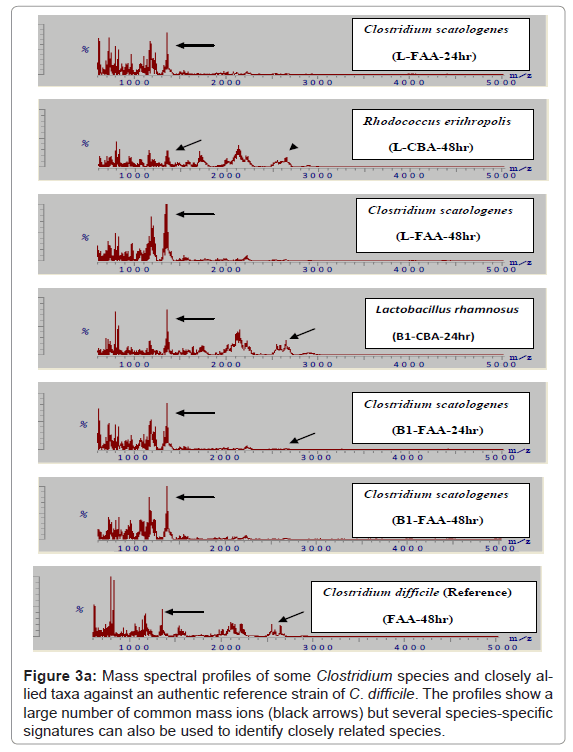
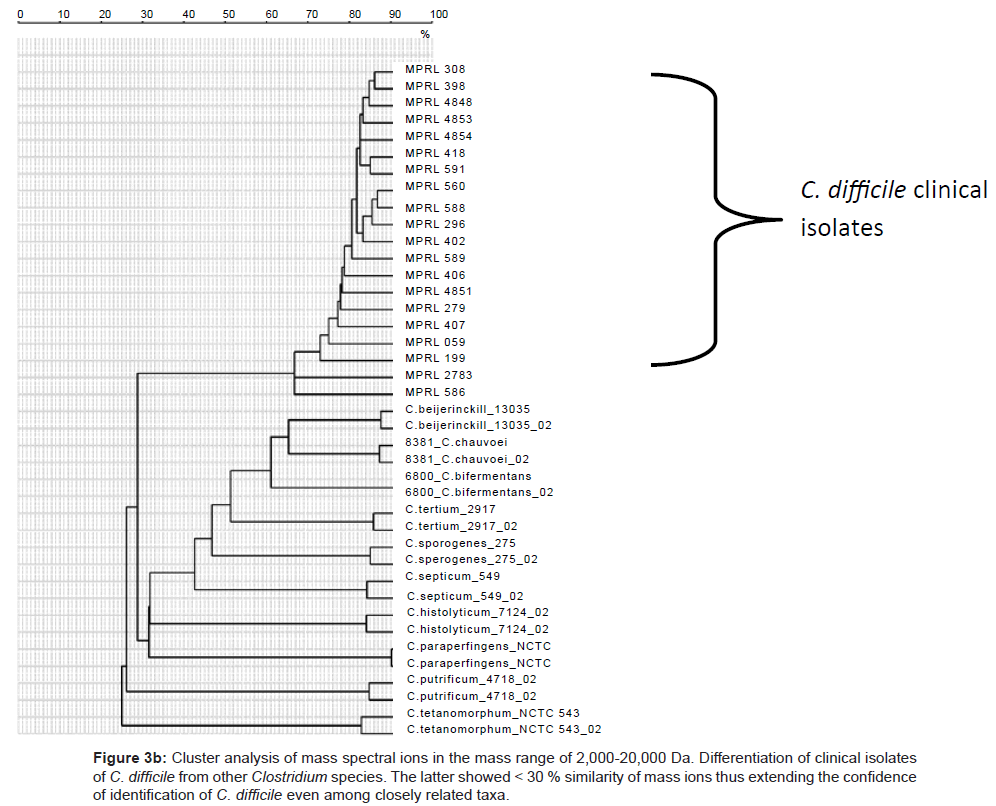
Because of the increased resolution and reproducibility of the spectra obtained with DHB, comparative analysis with other strains of C. difficile was then possible. Thus, from a total of 150 strains of this species all were shown to comprise comparable mass spectral profiles and clustered with a confidence of > 80 %. (Figure 2, Included as supplimentary figure) shows a dendrogram of this data and its separation from two other Gram positive species, P. acnes and Staphylococcus warneri and demonstrates unequivocally the utility of this approach for microbial diagnostic applications. Prior to this, only about 10% of C. difficile isolates were successfully identified [10]. Thus, despite the inherent diversity of this species, these results demonstrate there is a sufficient stable core of proteins that are retained to consolidate the isolates in the same phenon of the dendrogram. Mass spectra of several closely related clostridial species such as Clostridium bifermentans, Clostridium tertium, Clostridium septicum, Clostridium butyricum, Clostridium paraperfringens, Clostridium perfringens, Clostridium histolyticum and Clostridium tetanomorphum indicated that distinctive mass ions are produced that are characteristic of each species (Figure 3a) with species and generic markers being resolved that enables their differentiation as shown in Figure 3b.
Figure 3a: Mass spectral profiles of some Clostridium species and closely allied taxa against an authentic reference strain of C. difficile. The profiles show a large number of common mass ions (black arrows) but several species-specific signatures can also be used to identify closely related species.
Figure 3b: Cluster analysis of mass spectral ions in the mass range of 2,000-20,000 Da. Differentiation of clinical isolates of C. difficile from other Clostridium species. The latter showed < 30 % similarity of mass ions thus extending the confidence of identification of C. difficile even among closely related taxa.
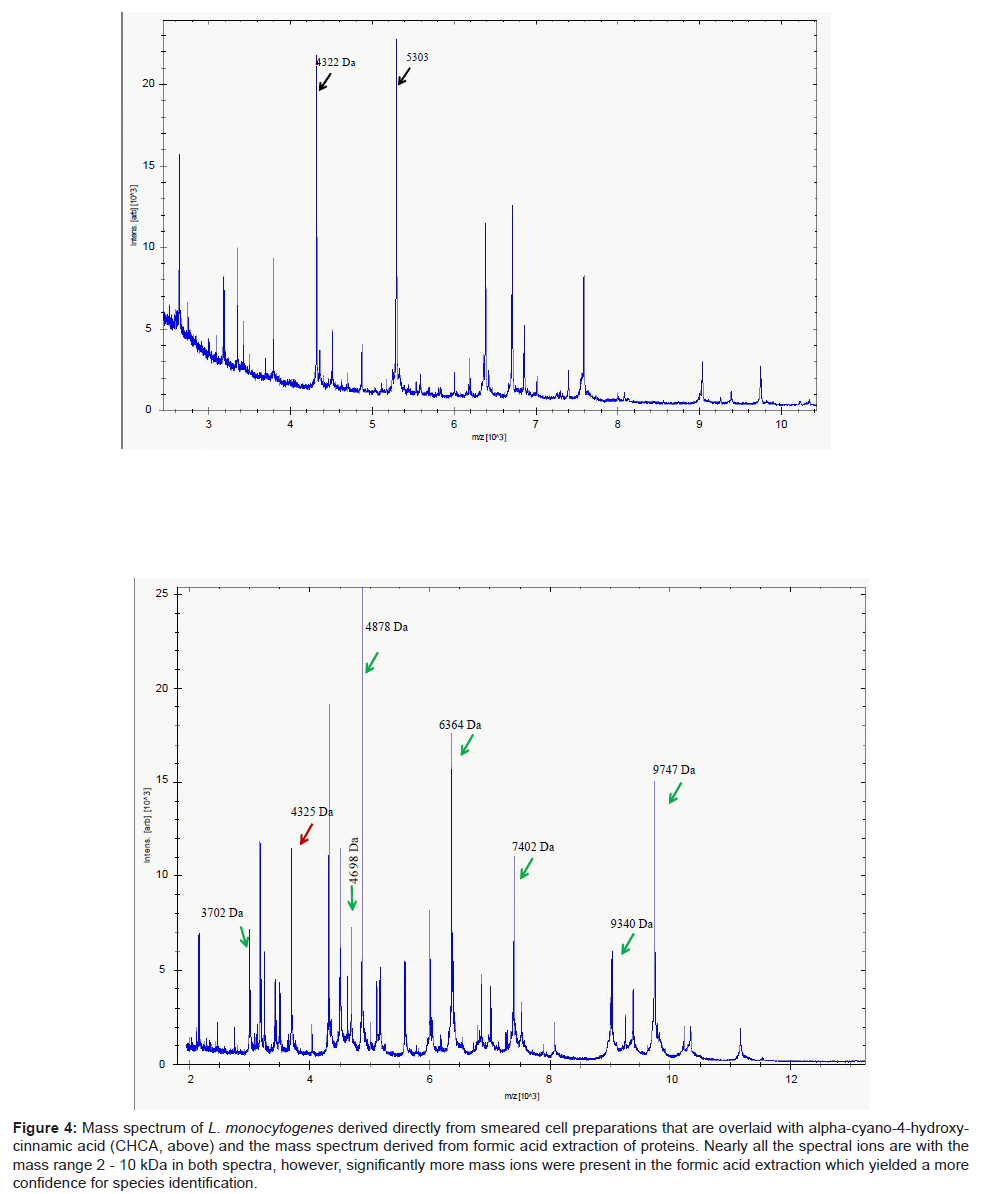
A parallel study with 13 strains of another Gram positive taxon, Listeria, revealed a similar pattern of results. Identification of isolates using DHB were considerably poorer compared to the use of CHCA. The mass spectra from the latter yielded a more compact profile of mass ions, most of which were prominent within the mass range 2-10 kDa (Figure 4). This permitted ready access to a database and therefore a high degree of confidence of identifying clinical isolates of Listeria species. In particular, the key pathogen of this genus, L. monocytogenes was readily delineated (data not shown). However, unlike Clostridium in which mass spectra were readily acquired by simply smearing the cells directly on to the target plate, mass spectra derived from formic acid extraction of proteins of Listeria species significantly improved the signal-to-noise ratio (Figure 4), improving the accuracy of analyte detection and consequently led to improvements and confidence of successful identification of these species.
Figure 4: Mass spectrum of L. monocytogenes derived directly from smeared cell preparations that are overlaid with alpha-cyano-4-hydroxycinnamic acid (CHCA, above) and the mass spectrum derived from formic acid extraction of proteins. Nearly all the spectral ions are with the mass range 2 - 10 kDa in both spectra, however, significantly more mass ions were present in the formic acid extraction which yielded a more confidence for species identification.
The general approach today for diagnostic microbiology is to compare mass ions in the range 2-20 kDa and for the majority of species most of the distinctive mass ions are packed into this mass range. This imposed restriction limits the resolution of the technique as a diagnostic platform. Thus, while MALDI-TOF MS is frequently cited as a high resolution tool for typing bacterial strains, for example for tracing disease outbreaks or for epidemiological studies, this is unlikely to be uniformly applicable across the microbial kingdom because of the limited number of strain-specific mass ions that can be reliably used to characterise an isolate. An alternative approach might be to extend the range of mass ions by selective capture of analytes using various chromatographic affinity devices and varying the matrix solution.
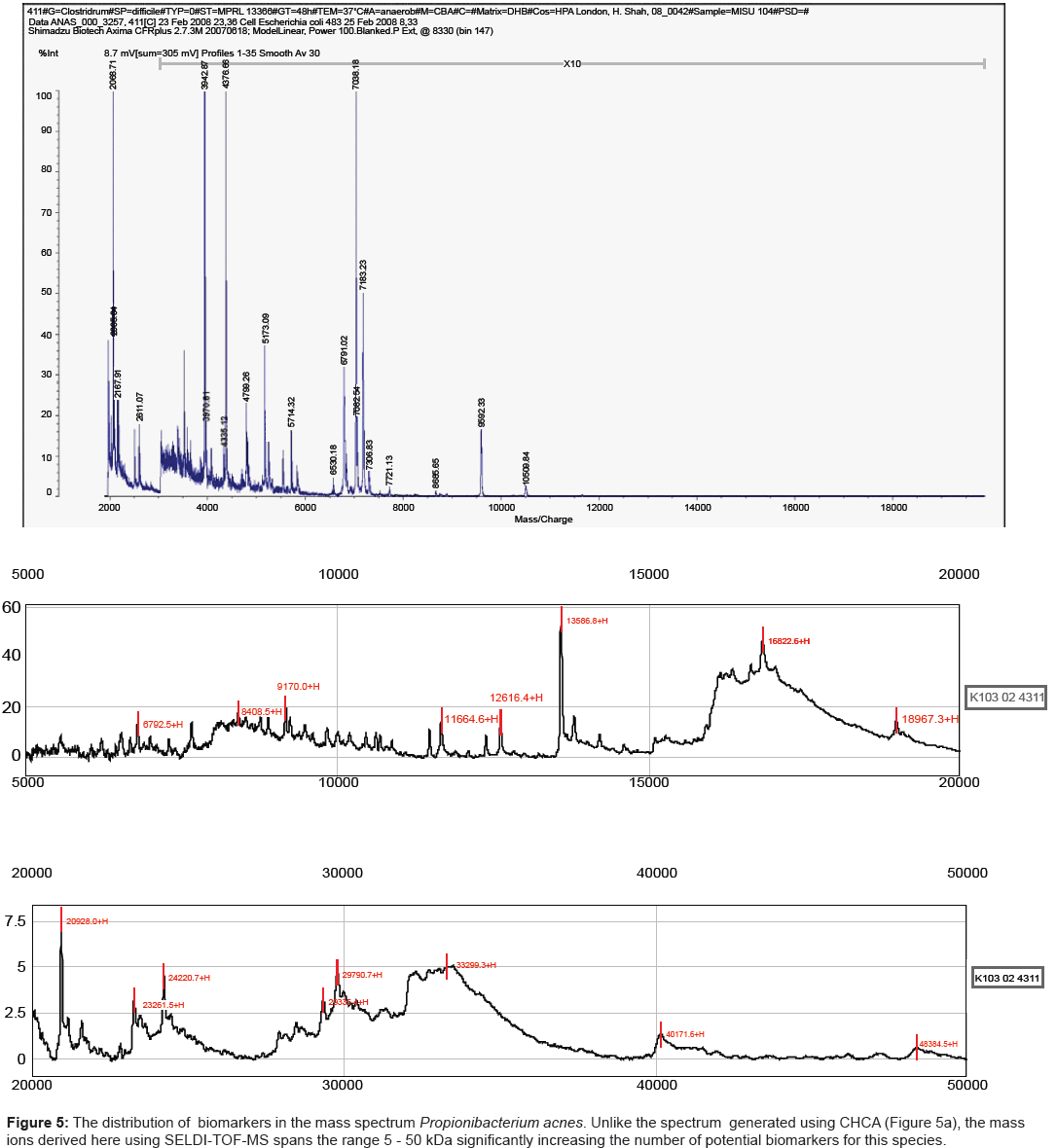
One such platform, referred to as SELDI-TOF MS, incorporates such parameters while still retaining the speed of analysis demanded for diagnostic applications. Potential analytes released by mechanical disruption of cells can be readily captured on an affinity chip, overlaid with matrix (sinapinic acid) and analysed by MALDI-TOF MS. We have shown previously that for a large number of species, the mass range is significantly increased, reaching up to 180 kDa for strains of Shigella species [25]. The results obtained here for strains of Propionibacterium acnes mirrored those reported previously for other species. Strains analysed using CHCA yielded spectra in which the major mass ions were less than 10 kDa (Figure 5a). By contrast the combination of using a hydrophobic capture chip with sinapinic acid as a matrix, extended the mass range by several orders of magnitude (Figure 5b).
Conclusions
The essential properties of MALDI-TOF MS matrices for high resolution mass spectral fingerprints are absorption wavelength, solubility, and crystal formation. Little is understood about the underlying physico-chemical processes that contribute to spectral information, however, it is widely accepted that the properties of the matrix solution is very much dependent on the sample-matrix solution composition, pH, and the rates at which the sample-matrix co-crystals are grown [26,27]. Table 1 summarises the relevant properties of the matrices used here for microbiological analysis and is in accord with the results obtained here for the three pathogens belonging to the genera Clostridium, Listeria and Propionibacterium. Thus, CHCA, CMBT and DHB all yielded mass spectra up to 10 kDA whereas SPA extended the mass range well beyond the level (Figures 5a and 5b).
The solvent composition of the matrix solution plays a key role in the process of co-crystallisation while the solubility of both matrix and peptides/proteins also affect the process. For example, high organic composition normally facilitates quick matrix crystallisation whereas low organic composition allows partial evaporation resulting in slow matrix crystallisation. The latter is believed to contribute to the suppressive effects on the matrix crystallisation process arising from high levels of non-volatile molecules that are frequently found in biological samples [26,27]. Hence, the pH and acid used to adjust the pH of the solvents used here, TFA or FA are expected to have a great impact on the number and intensity of the observed mass ions. Previous studies have indicated that TFA and FA and their concentrations have different effects on the mass spectra of intact bacterial cells. In general, TFA is a better ion pair agent than FA, it enhances the solubility of the proteins in the matrix, while FA is more favoured for its ability to increase the solubility of polypeptides [26-28].
Some bacterial taxa, particularly the more fragile Gram negative species readily lyse and release their proteins by the matrix solution. The more robust Gram positive species used here often need additional treatment prior to MALDI-TOF MS analysis. However, the results of this study showed that successful identification of these more robust Gram positive taxa was possible using directly smeared cells to which the matrix was overlaid as previously described [1,7]. Nevertheless, the results obtained here involving pre-extraction with 25% formic acid, which is known to increase the permeability of the cell wall and enhance the efficiency of protein extraction by the matrix solution, significantly improved the signal-to-noise ratio, improved the accuracy of analyte detection and led to improvements and confidence of successful identification of the test species.
References
- Keys CJ, Dare DJ, Sutton H, Wells G, Lunt M, et al. (2004) Compilation of a MALDI-TOF mass spectral database for the rapid screening and characterisation of bacteria implicated in human infectious diseases. Infect Genet Evol 4: 221-242.
- Honisch C, Chen Y, Mortimer C, Arnold C, Schmidt O, et al. (2007) Automated comparative sequence analysis by base-specific cleavage and mass spectrometry for nucleic acid-based microbial typing. Proc Natl Acad Sci U S A 104: 10649-10654.
- Kallow W, Erhard M, Shah HN, Raptakis E, Welker M, et al. (2010) MALDI-TOF MS for microbial identification: Years of experimental development to an established protocol. Mass Spectrometry for Microbial Proteomics. Wiley, Chichester, UK.
- Emonet S, Shah HN, Cherkaoui A, Schrenzel J (2010) Application and use of various mass spectrometry methods in clinical microbiology. Clin Microbiol Infect 16: 1604-1613.
- Justesen US, Holm A, Knudsen E, Andersen LB, Jensen TG, et al. (2011) Species identification of clinical isolates of anaerobic bacteria: a comparison of two matrix-assisted laser desorption/ionization–time of flight massspectrometry systems. J Clin Microbiol 49: 4314-4318.
- Claydon MA, Davey SN, Edwards-Jones V, Gordon DB (1996) The rapid identification of microorganisms using mass spectrometry. Nat Biotechnol 14: 1584-1586.
- Shah HN, Keys C, Gharbia SE, Ralphson K, Trundle F, et al. (2000) The application of Matrix-Assisted Laser Desorption/Ionisation Time of Flight Mass Spectrometry to profile the surface of intact bacterial cells. Microb Ecol Health Dis 12: 241-246.
- Shah HN, Keys CJ, Schmid O, Gharbia SE (2002) Matrix-Assisted Laser Desorption/Ionistaion Time of Flight Mass Spectrometry and Proteomics; a New Era in Anaerobic Microbiology. Clin Infect Dis 35: S58-64.
- Rajakaruna L, Hallas G, Molenaar L, Dare D, Sutton H, et al. (2009)High Throughput Identification of Clinical Isolates of Staphylococcus aureus using MALDI-TOF-MS of Intact Cells. Infect Genet Evol 9: 507-513.
- Rajakaruna L (2010) Proteomics as a tool for the characterisation of nosocomial pathogens. (Department for Bioanalysis and Horizon Technologies, Health Protection Agency Microbiology Services and Nottingham Trent University), UK.
- Vazquez-Boland JA, Dominguez-Bernal G, Gonzalez-Zorn B, Kreft J, Goebel W (2001) Pathogenicity islands and virulence evolution in Listeria. Microbes Infect 3: 571-584.
- Wang J, Yamada S, Ohashi E (2010) Rapid identification of Listeria species and screening for variants by melting curve and high-resolution melting curve analyses of the intergenic spacer region of the rRNA gene. Can J Microbiol 56: 676-682.
- Denny J, McLauchlin J (2008) Human Listeria monocytogenes infections in Europe - an opportunity for improved European surveillance. Euro Surveill 13: 8082.
- Nightingale K (2010) Listeria monocytogenes: knowledge gained through DNA sequence-based subtyping, implications, and future considerations. J AOAC Int 93: 1275-1286.
- Dekio I, Hayashi H, Sakamoto M, Kitahara M, Nishikawa T, et al. (2005) Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. J Med Microbiol 54: 1231-1238.
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, et al. (2009) Topographical and temporal diversity of the human skin microbiome. Science 324: 1190-1192.
- Dessinioti C, Katsambas AD (2010) The role of Propionibacterium acnes in acne pathogenesis: facts and controversies. Clin Dermatol 28: 2-7.
- Tunney MM, Patrick S, Curran MD, Ramage G, Hanna D, et al. (1999) Detection of prosthetic hip infection at revision arthroplasty by immunofluorescence microscopy and PCR amplification of the bacterial 16S rRNA gene. J Clin Microbiol 37: 3281-3290.
- Cohen RJ, Shannon BA, McNeal JE, Shannon T, Garrett KL (2005) Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: a possible link to cancer evolution? J Urol 173: 1969-1974.
- Lomholt HB, Kilian M (2010) Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS One 5: e12277.
- McDowell A, Gao A, Barnard E, Fink C, Murray P I, et al. (2011) A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology 157: 1990-2003.
- Dekio I, Rajendram D, Morita E, Gharbia S, Shah HN (2012) Genetic diversity of Propionibacterium acnesstrains isolated from human skin in Japan and comparison with their distribution in Europe. J Med Microbiol 61: 622-630.
- Higaki S, Kitagawa T, Kagoura M, Morohashi M, Yamagishi T (2000) Correlation between Propionibacterium acnes biotypes, lipase activity and rash degree in acne patients. J Dermatol 27: 519-522.
- McDowell A, Perry AL, Lambert PA, Patrick S (2008) A new phylogenetic group of Propionibacterium acnes. J Med Microbiol 57: 218-224.
- Shah HN, Encheva V, Schmid O, Nasir P, Culak RA, et al. (2006) Surface enhanced laser desorption/ionization time of flight mass spectrometry (SELDI-TOF-MS): a potentially powerful tool for rapid characterization of microorganisms. Encyclopedia of Rapid Microbiological Methods, DHI Publishing, USA.
- Williams TL, Andrzejewski D, Lay JO, Musser SM (2003) Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J Am Soc Mass Spectrom 14: 342-351.
- Cohen SL, Chait BT (1996) Influence of matrix solution conditions on the MALDI-MS analysis of peptides and proteins. Anal Chem 68: 31-37.
- Fenselau C, Demirev PA (2001) Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom Rev 20: 157-171.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14697
- [From(publication date):
specialissue-2014 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 10247
- PDF downloads : 4450