Research Article Open Access
Analyses using Cell Wall Glycan-directed Monoclonal Antibodies Reveal Xylan-degradation by Two Microbial Glycosyl Hydrolases in Cell Walls from Poplar and Switchgrass Biomass
| Supriya Ratnaparkhe1, Sivasankari Venkatachalam2, Michael G Hahn3 and Sivakumar Pattathil3* | |
| 1DBT-ICT-Centre for Energy Biosciences, Nathalal Parekh Marg, Matunga, Mumbai, India | |
| 2Department of Textiles, Merchandising and Interiors, University of Georgia, Athens, GA 30602, USA | |
| 3Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, USA | |
| Corresponding Author : | Sivakumar Pattathil Complex Carbohydrate Research Center University of Georgia, Athens, GA 30602, USA Tel: 706-542-4451 Fax: 706-542-4412 E-mail: siva@ccrc.uga.edu |
| Received: June 05, 2013; Accepted: September 06, 2013; Published: September 12, 2013 | |
| Citation: Ratnaparkhe S, Venkatachalam S, Hahn MG, Pattathil S (2013) Analyses using Cell Wall Glycan-directed Monoclonal Antibodies Reveal Xylan-degradation by Two Microbial Glycosyl Hydrolases in Cell Walls from Poplar and Switchgrass Biomass. J Bioremed Biodeg S4:004. doi:10.4172/2155-6199.S4-004 | |
| Copyright: © 2013 Ratnaparkhe S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Plant biomass represents the major source for renewable bio-fuels. Cell walls constitute the major portion of plant biomass. The main challenge in sustainable production of ligno-cellulosic biofuel is overcoming the cell wall recalcitrance barrier. A number of cell wall components including lignin and hemicelluloses have been shown to contribute to this cell wall recalcitrance. Thus, removing or reducing the proportion of wall components that contribute to cell wall recalcitrance of ligno-cellulosic biomass is a key step in sustainable ligno-cellulosic biofuel production. We have demonstrated the use of a comprehensive array of cell wall glycan-directed monoclonal antibodies to study the hydrolytic activities of two xylan-degrading microbial glycosyl hydrolases, CjXyl10B (a representative member of the GH10 family) and NpXyl11A (a representative member of the GH11 family) on cell wall extracts and cell walls from poplar and switchgrass biomass. Depletion of xylan epitopes, as monitored by the antibodies, in base extracts generated from raw poplar and switchgrass biomass that are treated with CjXyl10B or NpXyl11A confirmed the xylanase activity of these hydrolases, and demonstrates the utility of the antibody-based approach for screening for enzymes active on native biomass. Base extracts isolated from raw biomass treated with these enzymes prior to extraction also exhibited reduced xylan content. Further, prior removal of lignin significantly increased the efficiency of the xylan degradation in raw poplar and switchgrass biomass by the CjXyl10B and NpXyl11A enzymes. These results, thus hint that these two xylanases could potentially be used, in combination with lignin reduction, as efficient xylan-removing agents, while processing poplar and switchgrass feed stocks for biofuel production.
| Keywords |
| Monoclonal antibodies; Biofuels |
| Introduction |
| Plant biomass is considered as an important renewable resource for future bioenergy needs. Cell walls, which are typically highly recalcitrant, constitute the major part of this biomass. Overcoming the cell wall recalcitrance is considered as a major bottleneck towards achieving a highly sustainable and cost-effective production of biofuels from ligno-cellulosic materials [1]. Cell wall components, such as lignin and hemicelluloses, have been documented to contribute to cell wall recalcitrance [1,2]. In plant biomass, removal or reduction in the proportions of xylans, a major hemicellulose component of secondary wall-rich biomass, and that of lignin results in a significant reduction in cell wall recalcitrance [3,4]. This is because a reduction in xylan and lignin, in general, is thought to result in an increased exposure of cellulose microfibrils to subsequent microbial/enzymatic conversions [1]. Hence, any success in the generation of biomass raw materials with reduced amounts of hemicellulosic components, particularly xylan, could potentially be of high importance to efficient biofuel production from plant biomass. Such modifications are also important for other plant biomass-based bio-refinery applications, such as production of various value-added products including biochemical and biopolymers. |
| Hemicellulose is the second most abundant polysaccharide in biomass next to cellulose, and can represent up to 50% of total plant biomass. Based on the current state of knowledge, hemicelluloses can be divided into four general classes of structurally different cell-wall polysaccharide types, i.e. (a) xylans, (b) mannans (c) β-glucans with mixed linkages, and (d) xyloglucans. These polysaccharides include homo- and heteropolymers consisting largely of anhydro-β-(1-4)-Dxylopyranose, mannopyranose and glucopyranose main chains with a number of substituent’s, typically as side chains [5]. Hemicelluloses exhibit wide structural variations in the types of side-chains, their distribution, localization, and/or the types of glycosidic linkages in the backbone. Most research on hemicelluloses in the context of biofuels involves their use for conversion to sugars, chemicals, xylitol and feed stocks for the production of transportation fuels, such as ethanol and butanediol [6]. However, the structural diversity present in hemicelluloses also renders them attractive sources of biopolymers, which can be utilized in food or non-food applications. Enzymes that modify the structure of hemicelluloses can be exploited to obtain desired products from them, and hence, it is highly desirable to explore various ways to screen for and to characterize such enzymes. A large number of proteins thought to be glycosyl hydrolases on the basis of their amino acid sequences, together with their accessory carbohydrate binding modules (CBMs) have been grouped into sequence-based families in the Carbohydrate-Active EnZymes (CAZy) database [7]. However, only a very small percentage of these proteins have been subjected to detailed biochemical, structural or functional analysis, and hence, the activity of very few can be predicted correctly based on the available information. It is, therefore, important to conduct structural and biochemical analysis of these proteins in order to integrate the in silico information with actual experimental data [8]. |
| In the present study, we demonstrate a rapid and high-throughput method to investigate the hemicellulose-degrading properties of enzymes, in this case, belonging to CAZy families GH10 and GH11, and acting upon cell walls of poplar and switchgrass biomass. Xylans are the most abundant hemicellulose found in the plant cell walls of woody dicots and monocot grasses, and exhibit a heterogenous and complex structure [5]. Thus, the enzymes that can cleave xylans, e.g. xylanases, are likely to be quite diverse in their activities in terms of the structure recognized, enzymatic specificity and physico-chemical properties. Based on a search of databases, such as CAZy, the majority of xylanases cluster in the families GH10 and GH11 [9]. Both of these families encompass endoxylanases that hydrolyse β-(1,4)-linkages between adjacent xylopyranoside residues, except for a few GH10 xylanases that hydrolyse β-(1,3)-linkages [10], though structural differences distinguish the proteins in these two families. The GH10 family members have a larger average molecular mass of approximately 40 kDa, and their catalytic domain exhibits (β/α)8 architecture, with the active site being a shallow groove. On the other hand, the members of GH11 family have an average mass of approximately 20 kDa and display a β-jelly roll structure with a cleft shaped active site [11]. These structural differences have a bearing on their specificities. Enzymes of both families that have been characterized readily cleave arabinoxylans. GH11 xylanases preferentially cleave in unsubstituted regions of the arabinoxylan backbone, while GH10 xylanases are less hampered by 4-O-methyl-D-glucuronate, acetate and a-L-arabinofuranosyl decorations, along the xylan backbone. GH11 are the enzymes of choice in industries, as they are most active against insoluble polymeric xylans. GH10 xylanases, on the other hand, show a greater catalytic versatility, and can readily hydrolyse the products formed from the activity of GH11 enzymes, including small xylo-oligosaccharides, such as xylobiose and xylotriose [12]. |
| Immunological approaches have been used to some extent in combination with cell wall-degrading enzymes to study their effect on cell wall components [13-15]. However, to our knowledge, most of these studies utilized only a limited number of antibodies. Furthermore, immunological approaches to date have not been used to determine the activity of cell wall-degrading enzymes acting directly on biomass samples. Here, we demonstrate a high-throughput immunological screening method that can be used to ascertain information about the activities of proteins that might be active in cell wall degradation, focussing in this study on xylan degradation. We have used two representative enzymes for this study; a GH10, Cellvibrio japonicas xylanase CjXyl10B [16] and a GH11, Neocallimastix patriciarum xylanase NpXyl11A [17], and demonstrate that their activities on poplar and switchgrass biomass can be monitored by ELISA, using a large and comprehensive set of plant glycan-directed antibodies. We suggest that this technique can potentially also be used for rapid and high-throughput screening of uncharacterized glycan-degrading enzymes identified in databases, such as CAZy for their substrates and activity. In addition, we provide further evidence that this immunological approach can provide clues about interactions between various cell wall components. |
| Materials and Methods |
| Production of xylanases CjXyl10B and NpXyl11A |
| The xylanase CjXyl10B (a GH10 family enzyme) is from Cellvibrio japonicas and the xylanaseNpXyl11A (a GH11 family enzyme) is from Neocallimastix patriciarum. These enzymes were expressed as recombinant proteins in Escherichia coli. Gene construct construction, expression and subsequent enzyme purification were carried out as described [16-18]. Both recombinant proteins contain a C-terminal His-tag to facilitate purification. |
| Biomass samples |
| Biomass samples of poplar (Populus trichocarpa) and switchgrass (Panicum virgatum) that are 20 to 80 mesh particle size were provided through the BioEnergy Science Center, Oak Ridge National Laboratory (ORNL), and more details on the harvest and processing of these biomass samples are given elsewhere [3]. |
| Preparation of 1M and 4M KOH extracts |
| The 1 M and 4M KOH extracts from poplar and switchgrass were prepared in two different manners: |
| A) Alcohol insoluble residues (AIR) were prepared separately from poplar and switchgrass biomass [19]. The AIRs were extracted sequentially with 1 M and 4 M KOH, and the solubilized material processed as described earlier [19]. |
| B) The second set of samples was prepared directly from raw poplar and switchgrass biomass samples (Figure 1). First, the biomass samples were sequentially extracted with 1 M KOH and 4 M KOH to generate two control base extracts (extractions were done as explained earlier [19]). Another part of these raw biomass materials were initially treated with ammonium oxalate and sodium carbonate reagents, as described earlier [19], in order to remove pectic-polysaccharides that are extractable by these reagents. The resulting partially depectinated insoluble residues (500 mg) were suspended in 25 ml of 10 mM Tris- HCl buffer (pH 8.0) containing 150 Mm NaCl, and treated directly with CjXyl10B or NpXyl11A xylanases (10 μg/ml), separately at room temperature for 48 h. |
| The insoluble residues remaining after these initial enzyme treatments were again divided into two portions. One portion was sequentially extracted as above with 1 M KOH and 4 M KOH to generate two post-enzyme base extracts. The other portion of the enzyme-treated insoluble residues was used for lignin removal with sodium chlorite and glacial acetic acid treatment, as described [19]. These post-chlorite residues (delignified) were again divided equally into two portions and one portion was extracted with 4 M KOH to yield post-chlorite base extract controls. The second portion was treated with CjXyl10B or NpXyl11A overnight, as described above, and again extracted with 4 M KOH to yield post-chlorite enzyme-treated base extracts. A complete outline for the sequence of biomass treatments/ extractions explained above is shown in Figure 1. |
| All alkaline extracts were neutralized, dialyzed and lyophilized, as explained previously [19]. The total sugar content of each extract was determined using the phenol-sulfuric acid assay [20]. |
| In-plate digestion of 1 M and 4 M KOH AIR extracts |
| For the set of 1 M and 4 M KOH extracts that were prepared from AIRs of poplar and switchgrass biomass, 500 ng of each of the extracts were coated separately per well of a 96-well microtitre plate (Costar-3598) and dried overnight at 37°C. The immobilized extracts were then incubated overnight with 10 μg/ml of CjXyl10B or NpXyl11A xylanases (50 μl/well) at room temperature in 10 mM Tris-HCl buffer (pH 8.0), containing 150 mM NaCl. The wells were then washed three times with 10 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl. The enzyme-treated immobilized extracts were then subjected to enzyme-linked immunosorbent assays (ELISAs), as described below. |
| ELISA |
| All biomass extracts generated were subjected to ELISA with a collection of plant cell wall glycan-directed antibodies [19,21] (Supplemental Table 1), to analyze the overall glycan compositions with special emphasis on the xylan contents of these extracts. For the ELISAs, 500 ng (total sugar) of each extract were applied (in 50 μl) per well of 96-well plates (Costar-3598), and were allowed to dry down overnight at 37°C. The ELISAs were carried out essentially as described [19,21]. The plant cell wall glycan-directed monoclonal antibodies used in the current study were procured from laboratory stocks (CCRC, JIM and MAC series) at the Complex Carbohydrate Research Center (available through Carbo Source Services; http://www.carbosource. net), or were obtained from BioSupplies (Australia) (BG1, LAMP). Details of the McAbs used are given in the Supporting Information, Supplemental Table 1. |
| Results and Discussion |
| In general, removal of cell wall components, such as lignin and xylan from plant biomass, has been shown to contribute to a reduction in recalcitrance [3,4,22]. Identification and characterization of enzymes that can degrade cell wall components that contribute to biomass recalcitrance are an integral part of ligno-cellulosic bioenergy research. With an increasing amount of sequence information for carbohydratemodifying enzymes being added to the existing substantial databases of such enzyme sequences, it is imperative to develop rapid and high throughput methods for functionally characterizing the proteins encoded by these genes. Using immunological tools, we analysed the abilities of two microbial xylanases, CjXyl10B and NpXyl11A, from the glycoside hydrolase families GH10 and GH11, respectively, to degrade xylans from two prominent plant biomass feed stocks, poplar and switchgrass, targeted as biofuel feedstock crops. A large and diverse collection of plant cell wall glycan-directed antibodies were employed as a tool to screen for the glycan epitopes that are targeted by the two endoxylananses, as well as to monitor biomass samples for overall changes in wall structure arising from the action of these two enzymes. |
| In-plate digestion studies using base extracts from poplar and switchgrass cell walls |
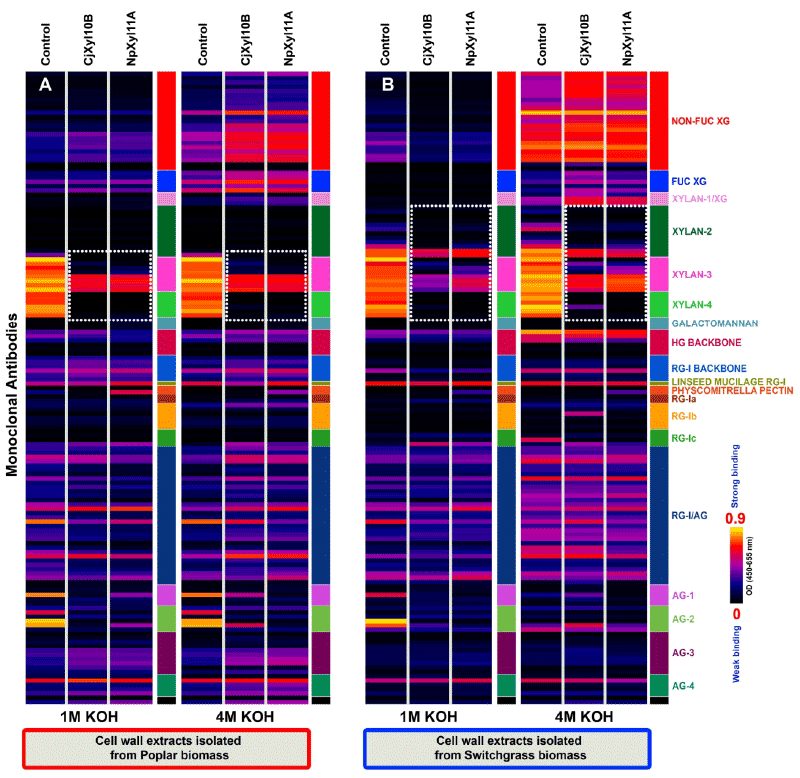
| Our primary targets for the two xylanases were xylan-rich cell wall extracts from poplar and switchgrass. Previous studies demonstrated that base (1 M KOH and 4 M KOH) extracts from both poplar and switchgrass cell wall preparations are enriched in hemicellulosic polysaccharides, especially xylans [3,22]. Hence, initial studies were conducted to test the abilities of CjXyl10B and NpXyl11A to breakdown extracted xylans after immobilization on microtiter plates, and to test the ability of xylan-directed antibodies to detect the action of these two enzymes on the immobilized substrates (Figure 2). Comparison of the enzyme-treated immobilized wall extracts with the controls showed that for poplar (Figure 2A), treatment with either CjXyl10B or NpXyl11A, resulted in the complete removal of all epitopes recognized by the xylan-4 group of antibodies. The xylan epitopes recognized by four xylan-3 McAbs, CCRC-M144, CCRC-M145, CCRC-M146 and CCRC-M155 (Figure 2A-white dotted blocks), appeared to be largely unaffected by treatment with either xylanase, whereas the epitopes recognized by the other McAbs in the xylan-3 group (CCRC-M137, CCRC-M149, CCRC-M152, CCRC-M160), are completely lost after treatment with the enzymes. For switchgrass cell wall extracts, the results of CjXyl10B and NpXyl11A action were similar, but clearly distinct from what was observed with the poplar extracts (Figure 2B). Unlike the control base extracts from poplar, switchgrass control base extracts contained epitopes recognized by several McAbs from the xylan-2 group, specifically CCRC-M113, CCRC-M114, CCRC-M116, CCRC-M117, CCRC-M150 and CCRC-M154 (Figure 2B). Such variations in the epitope composition in base extracts are indicative of variations in xylan structures and compositions between these two biomass types. With respect to the action of the two xylanases on switchgrass biomass, the xylan epitopes recognized by CCRC-M150 and CCRC-M154 were slightly reduced by CjXyl10B or NpXyl11A digestion (Figure 2B), while the epitopes recognized by the other four xylan-2 antibodies were lost after treatment with these enzymes. In addition, the epitopes recognized by four of the xylan-3 antibodies (CCRC-M144, CCRC-M145, CCRC-M146 and CCRC-M155) were only slightly affected by treatment with the two enzymes. The epitopes recognized by the other four xylan-3 antibodies (CCRC-M137, CCRC-M149, CCRC-M152, CCRC-M160) were largely, though not completely, lost by treatment of the switchgrass wall extracts with the two enzymes. The majority of the xylan epitopes destroyed by CjXyl10B or NpXyl11A treatment of wall extracts from both biomasses are those recognized by McAbs specific to unsubstituted homoxylan regions of xylans (xylan-4 group). |
| Interestingly, the action of the two enzymes on the xlyan epitopes bound by these latter xylan-3 antibodies were distinguishable from one another, based on the differing glycome profiles for these antibodies on the enzyme-treated wall extracts. These results suggest that there are subtle differences in the enzyme specificities of the two xylanases under study here, and also suggest that the antibody toolkit can be used to identify such activity differences. The exact nature of differences in enzyme specificity awaits detailed epitope characterization studies for the xylan-directed antibodies used in this study. These epitope studies are currently underway. |
| The relative insensitivity of xylan epitopes recognized by four xylan-4 antibodies (CCRC-M144, CCRC-M145, CCRC-M146 and CCRC-M155) in the base wall extracts from both biomass types and those recognized by two xylan-2 antibodies (CCRC-M150 and CCRC-M154) in switchgrass wall extracts to digestion by CjXyl10B or NpXyl11A demonstrates that the two xylanases studied here cannot entirely degrade all xylans present in these wall extracts. The resistance of these sets of xylan epitopes to xylanse digestibility could be explained in two ways. First, these enzyme-stable xylan epitopes in the base extracts are linked to other cell wall components, especially lignin, and hence are not accessible to these enzymes. Second, xylans in these biomass types occur as predominantly two classes, one having relatively few branches and the other being highly branched. The relatively unbranched xylans are cleaved by the xylanases into small fragments that no longer stick to the ELISA plate, and hence, the antibody signals for these epitopes disappear from the profiles. With the highly branched xylans, the xylanases may cleave and remove unbranched portions of the xylans, but leave behind fragments that are still large enough to be able to stick to the plate, and be recognized by xylan-directed McAbs that bind to branched or substituted xylans. |
| Another interesting aspect to the glycome profiling data for both the base extracts from enzyme-digested poplar walls, and for the 1 M KOH extracts from enzyme-digested switchgrass walls is that epitopes recognized by some AG-1 (JIM11) and AG-2 (MAC207, JIM12 and CCRC-M133) groups of McAbs were labile to both xylanase treatments. These results suggest that at least some arabinogalactans present in these extracts are linked to xylanase-digestible polymers in these biomass types. A recently reported study in Arabidopsis has documented a novel cell wall proteoglycan, arabinoxylan pectin arabinogalactan protein 1 (APAP1), that could account for such alink between pectic arabinogalactans and xylans [23]. If APAP1 orthologs exist in poplar and switchgrass, their presence could explain the loss of some arabinogalactan epitopes upon digestion of wall extracts with xylanases. |
| Other differences were also observed in the McAb binding patterns in the xylanase-treated base extracts from poplar and switchgrass cell walls. These include an increase in the binding intensities of xyloglycandirected McAbs (including antibodies recognizing non-fucosylated and fucosylated xyloglucan epitopes) to the 4 M KOH extracts from both wall samples treated with xylanases, and a subtle increase in the binding intensities of some of the AG-3 and most of the AG-4 groups of McAbsto, the xylanase-digested 4 M KOH extract from poplar walls. We cannot explain these changes to the glycome profiles of xylanasedigested wall extracts at the present time. |
| Studies using base extracts isolated from poplar and switchgrass biomass directly treated with xylanases prior to and postdelignification |
| The experiments above demonstrate that the xylanases can digest ELISA plate-immobilized xylans extracted from isolated cell walls of poplar and switchgrass. It was of interest to test the extent to which these xylanases can gain access to and digest xylans in intact biomass samples of both plants. We were also interested in determining whether or not delignification of the biomass samples would alter digestibility of the xylans in the biomass by the two xylanases. Such studies on the effectiveness of these two xylanases on raw biomass materials are specifically relevant to industrial applications for biomass processing and utilization. Additionally, studies of enzyme effects on raw biomass can provide insight into how xylans are integrated into the final wall structures of these two distinct biomasses, and how this integration varies between them. |
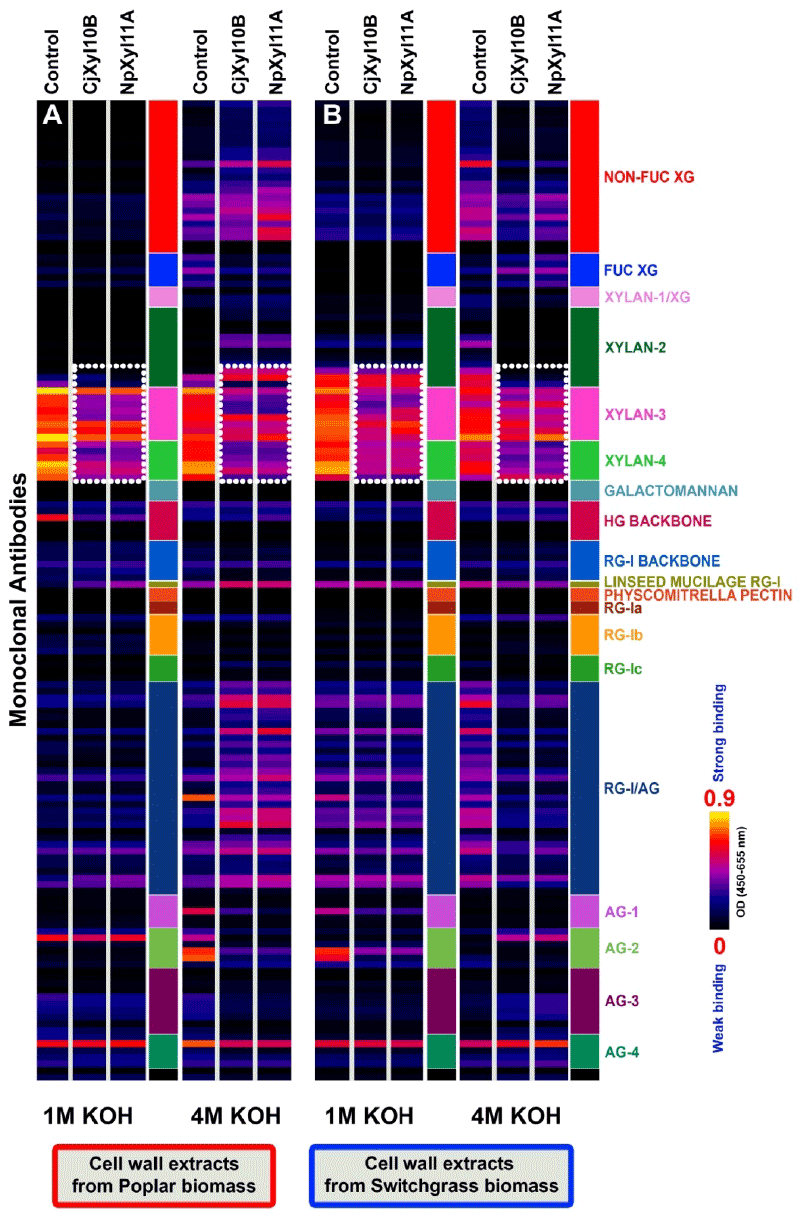
| The detailed experimental design for testing the effects of the two xylanases on raw and delignified biomass materials from poplar and switchgrass is shown in the scheme presented in Figure 1. The resulting wall extracts were screened against the antibody toolkit using ELISA, and the glycome profiles that resulted were largely identical in the case of both poplar (Figures 3A and 4A) and switchgrass (Figures 3B and 4B). These results suggest a common mode of action of both xylanases on these biomass types. In poplar and switchgrass, base extracts (1 M and 4 M KOHs) prepared from pre-delignified biomass residues (i.e. biomass that has been treated with oxalate and carbonate, as described in the Figure 1) that were treated with either CjXyl10B or NpXyl11A, resulted in a substantial reduction in the binding of xylan-3 and xylan-4 groups of McAbs, indicating a significant reduction in xylan epitope content in the biomass as compared to the respective controls (Figure 3A and 3B-white dotted blocks). These results shows that prior to delignification digestion with either CjXyl10B or NpXyl11A is not sufficient to completely remove all xylan epitopes in either poplar or switchgrass biomass. These results suggest that the xylans, even the unbranched domains that are susceptible to both enzymes (Figure 2) in these biomass residues, are not completely accessible to the digestive actions of the two xylanses. In pre-delignified poplar biomass, the digestion with both xylanases caused a significantly enhanced release of pectic-arabinogalactan epitopes into the 4 M KOH extract (Figure 3A). However, this trend was not observed in the case of pre-delignified switchgrass biomass, where the abundance of pectic-arabinogalactan epitopes was reduced in the corresponding 4 M KOH extract. These results suggest that in woody dicot biomass like poplar, cleaving xylans through xylanase treatments results in an increased extractability of pectin-arabinogalactans by high concentrations of base, suggesting some form of linkage exists in the wall between these polymers. An ortholog to the recently discovered proteoglycan, APAP1, which carries both xylan and arabinogalactan glycans [23], may constitute such a cross-link. |
| There were a couple of surprising results in the glycome profiles of xylanase-treated biomass. Digestion of pre-delignified switchgrass biomass with either CjXyl10B or NpXyl11A led to the complete removal of xylan-2 epitopes in comparison to the same extract from untreated controls (Figure 3B). Such a loss of these xylan-2 epitopes was not observed in the in-plate digestion studies with 4 M KOH extracts from switchgrass. These results suggest that there is a difference in accessibility of the xylan epitopes in muro compared with when the 4 M KOH base extracts from switchgrass were dried down on the ELISA plate. Another surprise was the presence, in the 4 M KOH fractions isolated from xylanase-digested pre-delignified poplar biomass, of xylan-2 epitopes (Figure 3A), since we had not observed xylan-2 epitopes in dicot wall extracts before. More research on this fraction will be required to explain this observation. |
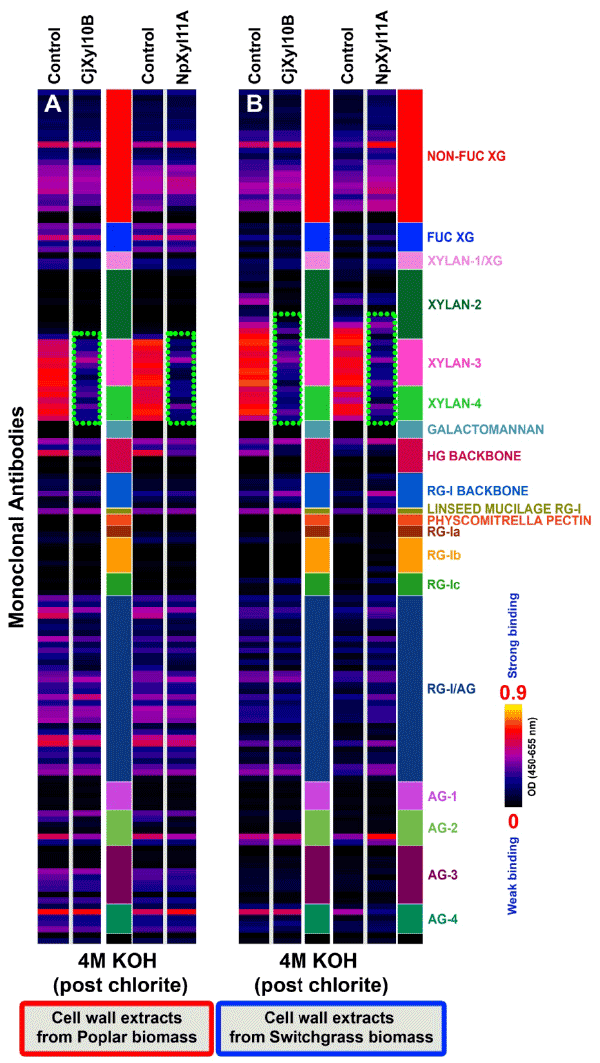
| It is generally accepted that in poplar and switchgrass, lignin interacts with hemicellulosic polysaccharides, especially xylans, and contributes to biomass recalcitrance [24,25]. Therefore, the xylanasetreated poplar and switchgrass biomass were subjected to chlorite treatment to remove up to 90% of the lignin (delignification), as demonstrated earlier in another plant biomass, black spruce [26]. Removal of the lignin resulted in an increased xylan extractability in a post-chlorite 4 M KOH extraction (see green-dotted boxes; Figure 4), compared with the pre-chlorite enzyme-treated biomass samples (Figure 3; white-dotted boxes). These results demonstrate that removal of lignin disrupted wall structure, in such a way as to render additional xylans extractable by base-treatment that were not extractable prior to delignification. Digestion of the chlorite-treated biomass with a second application of either xylanase resulted in the loss of almost all xylan epitopes from the 4 M KOH extract of treated biomass samples from both plants (Figure 4; green dotted boxes). These results indicate that removal of lignin significantly promotes accessibility of the CjXyl10B or NpXyl11A xylanases to their xylan substrates, thereby resulting in the almost complete removal of all classes of xylans from the two biomass samples. |
| Glycoside hydrolases are popularly used in the paper and bioethanol production industries, and are major contributors to the cost dynamics of these industries. A lot of studies are being carried out to obtain enzymes, and enzyme systems that have higher efficiency in degrading pretreated and/or untreated biomass. This volume of new research results in an increasing need to develop rapid and high-throughput methods that can monitor the functions of these enzymes/enzyme systems. It is also important that the substrate is made completely accessible to the enzyme, in order to tap its full potential. This can be achieved with better understanding of the interactions that exist between various cell wall components and eliminating the barriers that may exist in the wall that prevent or impair access of the enzymes to their substrates. The techniques demonstrated in this study can be applied to understand such interactions that exist amongst various biomass/cell wall components, and thus, assist in making their efficient hydrolysis achievable. |
| ELISA screening using cell wall glycan-directed McAbs: A promising rapid and high-throughput method for characterization of putative biomass-degrading enzymes |
| Putative glycosyl hydrolases are identified in large numbers and many of these proteins can potentially degrade broad classes of plant cell wall glycans. However, many of these proteins are assigned functions solely based on sequence similarities. Thus, high-throughput methods that can assist in functional characterization of these glycosyl hydrolases are in demand. The current study onCjXyl10B and NpXyl11A activities on poplar and switchgrass biomass shows that cell wall glycan directed McAbs based approaches provide an alternative to tedious characterization methods to conduct screening and short listing of substrates for cell wall deconstructing enzymes of unknown functions. The comprehensive suite of plant cell wall directed McAbs used in this study can also monitor most major non-xylan plant cell wall glycans. Hence, this toolkit can be used in a similar way to study the functions of enzymes that act on other wall polysaccharides. It should be noted that strong alkali extracts (especially 4 M KOH) isolated from plant biomass contain most major wall glycan components, including various hemicelluloses and pectins. Hence, this extract can act as a complex substrate mixture for testing the activities of diverse cell wall carbohydrate degrading enzymes of unknown functions. Taken together, in addition to the functional characterization of both xylanase activities on poplar and switchgrass biomass, these studies also demonstrate a high-throughput platform for functional analyses of carbohydrate active enzymes, that involves generating cell wall extracts with strong alkali, exposing these extracts to putative carbohydrate acting enzymes, and subsequently screening the enzyme-treated residues with glycan-directed McAbs to determine which polysaccharide components were affected by the action of the enzymes. |
| Acknowledgements |
| The authors wish to thank Prof. Harry Gilbert (University of Newcastle, UK) for the plasmids containing the CjXyl10B and NpXyl11A coding sequences. The glycome profiling was supported by the BioEnergy Science Center administered by Oak Ridge National Laboratory, and funded by a grant (DE-AC05-00OR22725) from the Office of Biological and Environmental Research, Office of Science, United States, Department of Energy. The generation of the CCRC series of plant cell wall glycan-directed monoclonal antibodies used in this work was supported by the NSF Plant Genome Program (DBI-0421683 and IOS-0923992). |
References
|
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14931
- [From(publication date):
specialissue-2014 - Apr 19, 2025] - Breakdown by view type
- HTML page views : 10227
- PDF downloads : 4704
