Commentary Open Access
Bioterrorism and Surveillance for Infectious Diseases - Lessons from Poliovirus and Enteric Virus Surveillance
L M Shulman1,2*, Y Manor1, D Sofer1 and E Mendelson1,2
1Central Virology Laboratory, Public Health Services, Israel Ministry of Health Sheba Medical Center, Tel Hashomer, Israel 52621
2Dept. of Epidemiology and Preventive Medicine, School of Public Health, Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel, 69978
- *Corresponding Author:
- Lester M. Shulman
Central Virology Laboratory
Chaim Sheba Medical Center
Tel-Hashomer, 52621, Israel
Tel: +972-3-530-2341
Fax: +972-3-535-0436
E-mail: lester.shulman@sheba.health.gov.il
Received Date: November 18, 2011; Accepted Date: February 05, 2012; Published Date: February 18, 2012
Citation: Shulman LM, Manor Y, Sofer D, Mendelson E (2012) Bioterrorism and Surveillance for Infectious Diseases - Lessons from Poliovirus and Enteric Virus Surveillance. J Bioterr Biodef S4:004. doi: 10.4172/2157-2526.S4-004
Copyright: © 2012 Shulman LM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioterrorism & Biodefense
Abstract
Early recognition and rapid response are crucial for control of infectious diseases introduced by bio-error, bio-terror or Mother Nature. Early recognition requires surveillance. Surveillance includes methods for identifying the presence of infectious agents or the symptoms caused by the presence of such agents. Overlapping of different surveillance strategies improves the chances for success. Results from enteric virus surveillance of acute viral gastroenteritis in sentinel children wards and outbreaks and environmental surveillance for polio and non-polio enteroviruses in Israel are presented to exemplify surveillance for infectious disease agents and for use as yardsticks for evaluating response to intervention and to introduction of new vaccination programs and for their potential for evaluating acute gastroenteris syndromic surveillance.
Introduction
Highly infectious pathogenic organism may be introduced into a region by bio-error, bio-terror, or Mother Nature. Bio-error is the accidental release of an infectious organism into the region. Examples include release from a vaccine production or storage facilities, archived biological material collected at the time when the organism was endemic, or from lab stocks either of the original organism or an extinct organism recreated by genetic engineering. Bio-terror is the intentional release of the infectious agent into the region from these sources. Finally, introduction by Mother Nature refers to the natural release of infectious organisms into a region. This most commonly occurs when infected hosts bring the organism into the region from an external reservoir or when changing living conditions increase the chance for zoonotic exposure within regions. Less common is reappearance as a result of release of an organism preserved in permafrost or by genetic evolution or genetic recombination in endogenous low pathogenic organisms that cause alterations in antigenicity reduce recognition by the immune system of individual hosts or host populations and/or which alter pathogenic organism-host interactions enabling the organism to change its host range or even the location within the host where that the organism can replicate. Included among low pathogenic organisms are live attenuated vaccines where evolution can lead to phenotypic reversion to a highly pathogenic form during infection of primary vaccines with functional immune systems, during persistent infections that may occur after vaccination or exposure of immune deficient individuals, and during subsequent person-to-person transmission to contacts of either of these types of individuals.
Early detection of the presence of the highly pathogenic infectious organisms is essential for containing the spread of these organisms in the flora or fauna within a given geographical region and for their elimination. This is true regardless of whether the infectious organism was introduced through bio-error, bio-terror, or Mother Nature. Early detection and measurement of efficacy of response requires high quality surveillance over extended periods of time. Integration of different surveillance strategies performed in parallel improves reliability and strengthens interpretation of data. The relatively unique combination of surveillance strategies for enteric viral infections in general and poliovirus infections in particular in Israel will be presented as a model for early detection of highly pathogenic organisms and subsequent monitoring of the efficacy of response. The main strategy that will be presented in detail uses environmental surveillance (reviewed in [1]) to identify the presence of pathogenic polioviruses in large populations before symptomatic cases occur. The second strategy that will be briefly reviewed is the systematic investigation of acute gastroenteritis in symptomatic cases from outbreaks and some or all admissions to sentinel departments in hospitals. Both approaches provide data about pathogenic organisms and provide a picture of what is going on at the community level. More importantly, both need to be in place for years to establish the base lines for recognizing the sudden appearance of unusual events. This depth is required to take into account seasonal and annual variations. Data gathered from these surveillance systems will also provide the basis for calibration and evaluation of a third approach, looking for unusual changes in the number and pattern of syndrome- specific admissions to hospital emergency wards or visits to HMO physicians. Specific examples will be provided to illustrate how classical and molecular data gathered from identification of pathogen-based strategies provide the epidemiological data needed for identification, response and follow up to bio-error, bio-terror, or Mother Nature triggered event.
Materials and Methods
Surveillance
Environmental surveillance for poliovirus and enteroviruses: Composite sewage samples have been routinely collected monthly from sentinel sites covering 30-40% of the Israeli population since 1989 using computerized automatic samplers that collect and pool aliquots hourly over a 24 hour period. When automatic sampling was not possible, peak hour grab samples were collected instead. Concentration, selection and isolation of polio and enteroviruses and downstream molecular epidemiological and phylogenetic characterization were as previously reported [2-5].
Acute viral gastroenteritis surveillance for enteric viruses: The enteric viral surveillance of severe acute gastroenteritis in sentinel children’s ward and outbreaks reviewed in the discussion was performed as published for rotavirus [6-8]. The norovirus was assayed by semiquantitative Real Time RT-PCR for the amplification and detection of norovirus genotype GII RNA by TaqMan technology as previously described [9].
Sydnromic surveillance for community physician visits for patients presenting with acute gastroenteritis from any cause: All Israelis are registered in one of four HMOs: Clalit, Maccabi Health Care Services, Kupat Holim Meuhedet, and Leumit. The HMOs maintain computerized records that include the ICD-9-based code reason for the visits. The Israel Center for Disease Control has received this data from the Maccabi Health Care Services, the second largest HMO covering approximately 25% of the population and Hospital Emergency Rooms on a daily basis for many years and prepares a weekly report on the rates of visits or admissions based on the ICD-9 codes. In this manuscript we present the weekly and monthly visits to the Maccabi Health Care Service community physicians of patients who presented between January 2004 and June 2008 with acute gastroenteritis from any cause [protozoal intestinal disease uns, bacterial enteritis uns, viral enteritis uns, enteritis uns, infectious colitis, acute gastroenteritis, proven gastroenteritis infection, and presumed gastroenteritis infection] and for nausea and vomiting from any cause.
Electron microscopy: 10 μl of clarified stool suspension was dried on Smart grids (Dune Sciences, Or, USA). The material on the grids was negatively stained with uranyl acetate and then viewed on a Jeol LS200 EX II transmission electron microscope (Jeol LTD, UK) at a magnification of 30,000 to 40,000. Final identification on the basis of size and morphology was made at a magnification of 100,000.
VP1 Sequences: The VP1 sequences of the type 1 WP isolates represented in the phylogenetic tree in Figure 1 were submitted to the EMBL/GenBank/DDJB data bank. The accession numbers are listed in parenthesis after the short name (as it appears in Figure 1) and the full isolate name. Isolates from the Gaza District: P-1 = PV1/2335_1/ PAL91 (JQ228553); P-2 = PV1/2252_1/PAL91 (P2 JQ228554); P-3 = PV1/3313_25/PAL94 (JQ228555); P-4 = PV1/3313_27/PAL94 (JQ228556); P-5 = PV1/3380_7/PAL94 (JQ228557); P-6 = PV1/3381_3/ PAL94 (JQ228558); P-7 = PV1/3381_9/PAL94 (JQ228559); P-8 = PV1/3432_5/PAL95 (JQ228560); P-9 = PV1/3431_21/PAL95 (JQ228561); P-10 = PV1/3431_21/PAL95 (JQ228562); and P-11= PV1/3455_27/PAL95 (JQ228563). Israeli isolates: Is-1 = ISR87-5483 (AF528790) and Is-2 = PV1/3615/ISR95 (JQ228564).
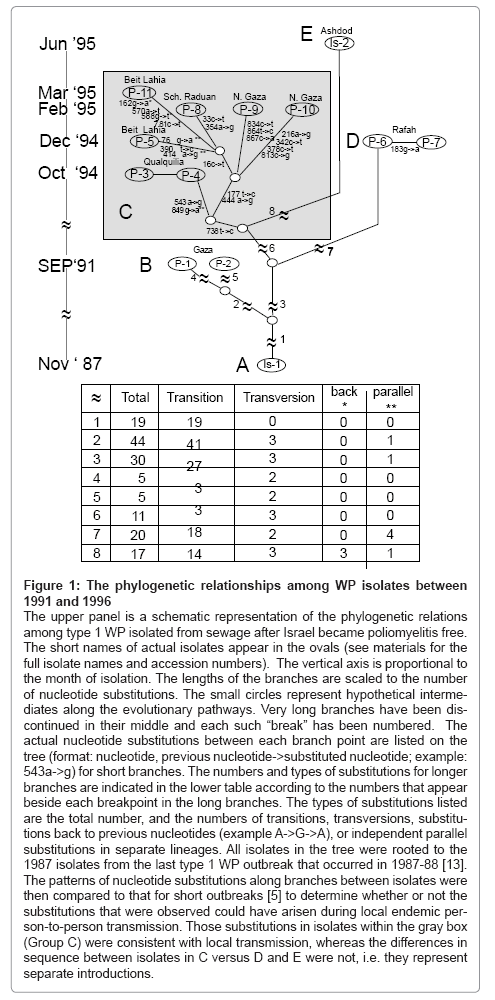
Figure 1: The phylogenetic relationships among WP isolates between 1991 and 1996
The upper panel is a schematic representation of the phylogenetic relations among type 1 WP isolated from sewage after Israel became poliomyelitis free. The short names of actual isolates appear in the ovals (see materials for the full isolate names and accession numbers). The vertical axis is proportional to the month of isolation. The lengths of the branches are scaled to the number of nucleotide substitutions. The small circles represent hypothetical intermediates along the evolutionary pathways. Very long branches have been discontinued in their middle and each such “break” has been numbered. The actual nucleotide substitutions between each branch point are listed on the tree (format: nucleotide, previous nucleotide->substituted nucleotide; example: 543a->g) for short branches. The numbers and types of substitutions for longer branches are indicated in the lower table according to the numbers that appear beside each breakpoint in the long branches. The types of substitutions listed are the total number, and the numbers of transitions, transversions, substitutions back to previous nucleotides (example A->G->A), or independent parallel substitutions in separate lineages. All isolates in the tree were rooted to the 1987 isolates from the last type 1 WP outbreak that occurred in 1987-88 [13]. The patterns of nucleotide substitutions along branches between isolates were then compared to that for short outbreaks [5] to determine whether or not the substitutions that were observed could have arisen during local endemic person- to-person transmission. Those substitutions in isolates within the gray box (Group C) were consistent with local transmission, whereas the differences in sequence between isolates in C versus D and E were not, i.e. they represent separate introductions.
Results and Discussion
Environmental surveillance for polioviruses
Symptomatic and asymptomatic individuals excrete virus progeny into sewage during enteric viral infections. This is the basis behind the use of environmental surveillance to document poliovirus infections in populations [10]. More importantly from the public health point of view, sewage surveillance can detect silent poliovirus virus circulation before appearance of the first case of irreversible poliomyelitis [1] since less than 1% of infections of naïve children even with highly pathogenic wild poliovirus cause poliomyelitis, whereas >90% are asymptomatic. Most surveillance procedures involve concentrating the virus in a sewage sample followed by a biological amplification of the viruses on tissue culture before molecular analysis [1,11]. Countrywide routine monthly environmental surveillance for poliovirus in sewage was initiated in Israel in 1989 and is the longest running national comprehensive surveillance program to date [1]. Approximately 30- 40% of the entire population lives within the catchment areas of the surveillance sites. Since the initiation of surveillance, tourists and businessmen have traveled to and from regions were polio was still endemic. In addition there was a large influx of Palestinian families from poliovirus endemic regions into the West Bank and Gaza District in 1994-1995 after the signing of the Oslo Peace accords.
When polioviruses replicate, they have very high rates of nucleotide miss-incorporation because their RNA primed RNA polymerase lacks a proofreading capability [12]. The genomic regions encoding the entire VP1capsid protein from isolates from the last outbreak of poliomyelitis in Israel that occurred in 1987-1988 [13,14] were sequenced to establish the first high-resolution, full VP1 gene, molecular analysis of the rate and pattern of nucleotide changes that occur during short outbreaks arising from a single founder [5]. Jorba et. al. [12] analyzed additional outbreaks, some extending over many years, to establish a series of rates suitable for characterizing the relatedness of polioviruses isolated at very short to very long intervals depending on the classes of substitutions measured. These classes include synonymous third codon position substitutions, non-synonymous substitutions and transversion. These rates and the 10:1 ratio of transitions to transversions [12] can be used as a benchmark to distinguish between the possibility that two polioviruses isolated within a short time interval arose during personto- person transmission during an outbreak or resulted from two separate introductions from an external reservoir or different external reservoirs [2]. Introduction from an external reservoir is further supported when the pair-wise comparison of nucleotide sequence homology among isolates in the surveillance area is lower than between some of these isolates and isolates from external reservoirs [2].
Based on the performance of long-term environmental poliovirus surveillance (reviewed in [1]), the WHO has recently recommended that routine sewage surveillance be established in more countries (17th Informal Consultation on the Global Polio Laboratory Network, 2011, Geneva) based on standards previously recommended for environmental surveillance for polioviruses [11]. All of the recommended methods required a biological amplification and selection step. Specifically growing poliovirus in tissue cultures and using transgenic L20B marine cells. L20B cells express the human receptor for polioviruses and selectively amplify poliovirus but not other human non-polio enteroviruses [15,16]. Many of the countries that have or will institute environmental surveillance are wild poliovirus free and most or all samples should be negative for polioviruses. Sample integrity is especially important for the interpretation of a negative isolation. It was therefore important to introduce a control to ensure that the quality and integrity of the sample was maintained from the time of collection, during transport, and during processing in the laboratory. The control that was recommended [11] was to also test for the presence of nonpolio enteroviruses using cell lines that support growth of most human enteroviruses. The reasoning behind such a control is that the lability of other enteroviruses is similar to that of polioviruses and enterovirus infections are common everywhere.
The type of poliovirus in sewage depends in part on the vaccination program in use. There are three types of poliovirus: vaccine, wild (WP) and vaccine-derived poliovirus (VDPV) [17]. An isolate was defined as a vaccine strain if the difference between the nucleotide sequence encoding its VP1 capsid protein and its respective Sabin serotype was <1%. It was considered to be a VDPV, when its VP1 sequence divergence ranged between 1% and 15%. Isolates with >15% divergence were considered to be WP. Recently the upper limit for VDPVs was modified to include isolates with >15% divergence that could be phylogenetically related to VDPVs with <15% divergence. This change was recently instituted (Summary of the 16th Informal Consultation on the Global Polio Laboratory Network, Geneva, Switzerland, 2010) after it was shown that a number of isolates with >15% divergence were genotypically related to previously isolated VDPVs from long standing environmental surveillance programs carried out by Israel (discussed below) and Finland (reviewed by Hovi et al [1]).
Isolation of vaccine virus
Between 1989 and 2005, all children in Israel were vaccinated by the age of 15 months with three doses of inactivated polio vaccine (IPV) and three doses of live attenuated Sabin vaccine (OPV) [13]. From 1990 they were also vaccinated with an additional IPV dose at 6-7 years of age [13]. Annual vaccine coverage of at least three doses exceeded 95%. During the period when OPV was included in early childhood vaccination schedules, most of the live vaccine virus actually ended up in sanitary dumps because of the use of diapers, however some especially from older contacts routinely entered the sewage system. During this time >95% of the polioviruses isolated from sewage were vaccine strains. After 2005 Israel switched to exclusive use of IPV. OPV strains then rapidly disappeared from the sewage as had occurred in other countries that made a similar shift to exclusive use of IPV in their vaccination policies [1].
Isolation of WP
The appearance of wild poliovirus isolates in sewage collected from a poliomyelitis free region indicates the silent presence or transmission of wild virus. The longer the poliomyelitis free period, the more likely that the WP has been introduced from an external reservoir. No specific intervention is required when there is herd immunity sufficient to prevent local transmission, for example when documented vaccine coverage is >95%. Rapid intervention (vaccination) is required, however, when coverage levels are below this level or when there is evidence of local circulation.
Wild type 1 polioviruses were recovered from sewage collected from various sites in the Gaza District in 1991, 1994, 1995, 1996, 1999 and 1992 and from Ashdod, Israel in 1995. The entire VP1 of representative isolates were sequenced. The phylogenetic relationships among WP isolates collected between 1991 and 1996 are shown in Figure 1. The time course and pattern of nucleotide substitutions among the WP isolates was compared to the pattern from the 1987-1988 outbreaks [5]. The number of nucleotide differences between isolates in groups C (1994-5), D (1994) and E (1995) was much higher than the 1% per year expected for isolates from the same outbreak [2,12]. This large deviation from the expected rate for nucleotide substitutions and somewhat smaller deviations from the 10:1 transition to transversion ratios allowed us to infer that the isolates in group C, D and E in Figure 1 were progeny from separate introductions. In contrast, the kinetic pattern among group C isolates was consistent with the pattern of local transmission that occurred during the outbreak in 1987-1988 [5]. Furthermore, these WP isolates appeared during the peak in the influx of whole families with unknown vaccination histories into the Gaza District. This combination of events triggered an immunization response. Sewer surveillance was able to document the efficacy of the immunization response [1,4]. Specifically, there was a rapid decrease and disappearance of genotype-related WP isolates from subsequent samples from these positive sites.
In 2002, two WP isolates were recovered within a four-month interval from the Gaza District. Using the same approach for analysis, these were considered to be from separate introductions [2]. Namely there was a much higher difference in nucleotide substitutions than could be expected to occur within four months for an outbreak with a single founder, the transition transversion ratios were inconsistent, and the relative homology indicated that each isolate was more homologous with isolates from different regions of Egypt than the Gaza District isolates were to each other.
Two important principles that apply to bio-defense are illustrated from these WP surveillance studies. Advanced molecular analysis can indicate when virus isolates are introduced into a virus-free region, can infer whether such isolates represent subsequent local circulation or separate introductions, and can identify their probable external reservoir. This in turn can trigger the type of response in the surveillance region, and the same surveillance program can then monitor the efficacy of the response. A further advantage is that response can also be extended to the region of the external reservoir.
Isolation of VDPVs
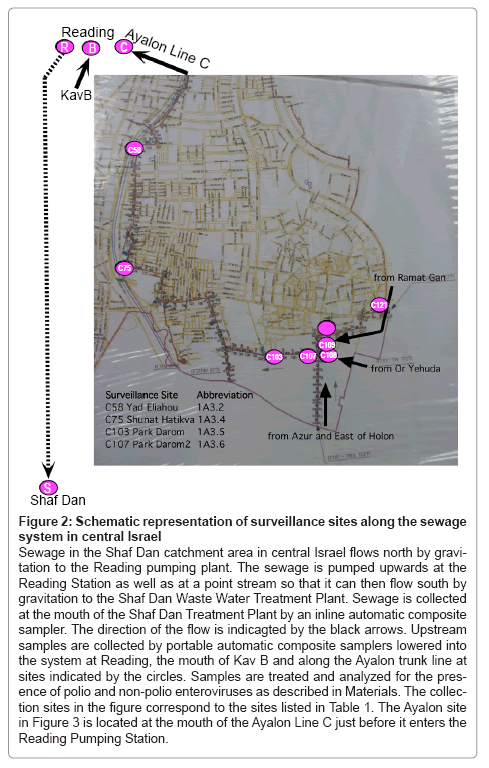
In May of 1998, a serotype two VDPV (VDPV2) was isolated from a sewage sample collected at the entrance to the Shaf Dan wastewater treatment plant in central Israel [18]. Surveillance sites that were subsequently introduced throughout the catchment area of this sewage system are shown in Figure 2. In 1998, the catchment area of the Shaf Dan Plant encompassed a population of 1,600,000 individuals. Sequence analysis of the VP1 of the 1998 VDPV2 indicated that it had diverged by 8% from Sabin 2 and that extensive amino acid substitutions were present in neutralizing antigenic epitopes of the capsid proteins.
Figure 2: Schematic representation of surveillance sites along the sewage system in central Israel
Sewage in the Shaf Dan catchment area in central Israel flows north by gravitation to the Reading pumping plant. The sewage is pumped upwards at the Reading Station as well as at a point stream so that it can then flow south by gravitation to the Shaf Dan Waste Water Treatment Plant. Sewage is collected at the mouth of the Shaf Dan Treatment Plant by an inline automatic composite sampler. The direction of the flow is indicagted by the black arrows. Upstream samples are collected by portable automatic composite samplers lowered into the system at Reading, the mouth of Kav B and along the Ayalon trunk line at sites indicated by the circles. Samples are treated and analyzed for the pres- ence of polio and non-polio enteroviruses as described in Materials. The collec- tion sites in the figure correspond to the sites listed in Table 1. The Ayalon site in Figure 3 is located at the mouth of the Ayalon Line C just before it enters the Reading Pumping Station.
Between May 1998 and Sept 2011, 62 phenotypically related VDPV2s were intermittently isolated from an additional 43 sewage samples. During this period of surveillance, two strategies were pursued to increase isolations: sampling frequency was increased at established sites and additional sites were successively added at points above positive sites where major trunk lines converged (see Figure 1). Sewage samples that contained VDPV2s are shown by collection date in Table 1. Colored squares (pink, blue, and yellow) indicate a site that was positive for VDPV, white boxes indicate negative sites that were sampled within a month of the positive site, while grey boxes indicate a negative finding at a site that was sampled on the same day as the positive site. From left to right, columns in Table 1 represent the progressive spatial transition from the mouth of the Shaf Dan treatment plant to the most upstream site (see Figure 2). The rightmost two columns represent a positive VDPV2 site from Jerusalem and the only site from where a type 1 VDPV was isolated, a site in Haifa.
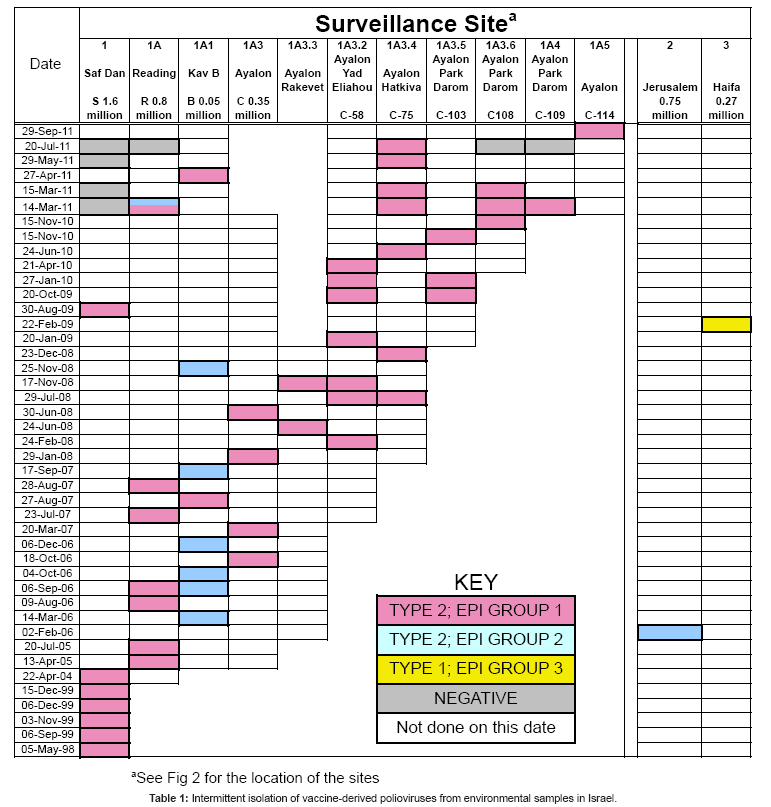
After a half-year trial, surveillance schedules were returned to previous frequencies since the increase in sampling frequency added considerably to the workload without increasing the frequency of recovery. In contrast, adding upstream surveillance sites proved effective in increasing isolations. Contributing to this was that VDPV2 virus from the excretor progressively increased as the site of excretion was approached. The catchment population of the first major positive junction site, Redding, was reduced to 800,000 and successive addition of upstream sites along the Ayalon trunk line, one of five that fed into the Reading site, have further reduced the population including the excretor to approximately 25-50,000 individuals.
The VP1 of each isolate was sequenced. Samples that contained VDPV2 isolates that were phenotypically related to the 1998 VDPV2 are represented by the pink boxes in Table 1. On Feb 2006, a phenotypically unrelated VDPV2 with 6.6% VP1 divergence was recovered from a single collection in Jerusalem. No related isolates have been found to date from that site in Jerusalem, however by Sept 2011, eleven phenotypically related isolates (represented by blue boxes in Table 1) have been recovered from eight samples from the Shaf Dan catchment area in Tel Aviv, starting with one from the Redding site that was isolated one month after the isolation in Jerusalem. The next ten related isolates were recovered from the Redding site or trunk line, Kav B, which is a different upstream trunk line from the sites that were positive for isolates related to the 1998 VDPV2. The catchment area of Kav B includes most of the major Mediterranean Sea shorefront hotels in north Tel Aviv and represents a 50,000 population. A change in the sewage system but not surveillance site has since reduced the catchment population for the Kav B positive site to 25,000.
The Shaf Dan-like isolates (pink boxes in Table 1) and the Jerusalem-Redding like VDPV isolates (blue boxes in Table 1) belong to two separate epidemiological events, i.e. persistent infection of two different immune individuals after separate exposures to serotype two Sabin vaccine [19]. The nucleotide substitutions within the VP1 genes from the VDPV2 isolates steadily increase from 8% to16.7% as time progressed. The VPDV2 isolates were shown to be highly neurovirulent in mice model systems and neutralizing antibody titers in the general public were three-fold lower than against respective Sabin strains [3,19]. Fortunately, despite the high nucleotide and amino acid divergence, these isolates were still as sensitive in vitro as the Sabin 2 strain to the antiviral effects of isoflavenes [20].
The identity of the individuals excreting the aVDPVs has not yet been established. To infer more about the excretors, additional segments of the 7400 nt genome of each isolate were sequenced. These additional regions included the 5’untranslated region and VP2, VP3 and VP4 capsid protein genes located 5’ of the VP1 capsid gene, and the RNA primed RNA polymerase, 3D, located at the 3’ end of the single open reading frame of the poliovirus genome. Phylogenetic analysis of all of the environmental aVDPV isolates indicated that excretion was limited to one individual and/or a very limited number of contacts for each of the epidemiological events. In the absence of identification of the excretor or excretors, this conclusion was based on the following lines of reasoning. (1) Most nucleotide substitutions that accumulate during evolution of poliovirus are random and most are silent third codon position substitutions [12,21,22]. Two different patterns involving high numbers of identical substitutions spread throughout the genome, despite overall sequence diversion of >10% among isolates indicated that there were two, and only two epidemiological events [19]. (2) Progeny that arose during person-toperson transmission in populations of immune competent individuals, cVDPVs, have very few amino acid substitutions in antigenic sites and most rapidly undergo multiple recombination with other polioviruses and non-polio enteroviruses [21,22]. In contrast, progeny that arise during persistent infections in immune deficient individuals have many amino acid changes in antigenic epitopes and most had not undergone any genomic recombination [21,22]. The Israeli aVDPV isolates were inferred to have come from persistent infections of either of two immune deficient individuals since the aVDPVs from each event had many amino acid substitutions in antigenic sites and there was different single recombination event within the polymerase gene for isolates of each event [19]. (3) Immune deficient persistently poliovirus infected individuals are rare; only 40 identified throughout the world to date [17]. (4) Vaccine excretion in healthy individuals is self-limiting and measured in weeks at most [23,24]. During the twelve-year interval of aVDPV isolations, the annual polio vaccine coverage of the Israeli population was >95% [13]. Thus the conditions for establishment of herd immunity were in place meaning that there would have been an insufficient number of naïve immune competent individuals available to maintain a chain of transmission over the 12 years during which the excretor or excretors were present in the catchment area, or the 24 years during which the virus evolved. And (5) the consistent recovery of each group of event-related isolates at separate geographical locations within the same city, e.g., at surveillance sites along different branches of the sewage system in a single city, when 30-40% of the entire country-wide population has been subjected to monthly surveillance throughout this period argues for minimal person-to-person spread. These arguments rely on one of the basic tenets of a good surveillance program, namely that in order to discover or rule out occurrence of an unusual event, there must be sufficient data from long-term routine surveillance.
Downstream sites are frequently negative when upstream sites are positive, and detection is intermittent even at the most upstream sites. This is shown by the surveillance entries for March 14th and 15th , May 29th , and July 20th of 2001 (Table 1) where multiple sites (see Figure 2) were simultaneously monitored at hourly intervals by automatic samplers; three sites for two successive 24 hour period and two sites for just the first 24 hour period. At four of the sites, composite samples were obtained for each 24- hour period by pooling samples. At the 5th site (C-75), hourly samples were collected and analyzed separately. On March 14th, starting from downstream sites and proceeding to upstream sites, no VDPVs were recovered from the Shaf Dan site, two VDPV2s were isolated from the Reading site, three of the first set of 24 hourly samples from C-75 located upstream contained a single VDPV2, two VDPV2s were isolated from site C-108 and one from C-109. On March 15th, no VDPV2s were recovered from the Shaf Dan site, one VDPV2 was recovered from one of the 24 individual samples from C-25 and one from site C-108. All of the isolates from the Ayalon line sites (C-75, C108, and C109) were related to previous Shaf Dan-Ayalon isolates. One of the two Reading VDPV2 isolates was related to these Shaf Dan-Ayalon VDPVs, while the other was related to the Jerusalem-Kav B isolates. The isolates from C-75 were obtained from samples collected at 09:00, 10:00, 04:00 and 08:00 AM suggesting that the virus is excreted from a residential site or less likely from a nighttime job.
These VDPV studies illustrate four additional principles and some limitations of environmental surveillance [1] that can be applied toward bio-defense. (1) Environmental surveillance is sensitive enough to detect virus excreted by one or a very small number of individuals in very large populations. (2) A negative finding does not mean the absence of the virus since the sensitivity is at the limits of detection. Most virus positive sewage samples contained one or at most a few poliovirus isolates, whereas it is estimated that 107 viruses are excreted per gram of feces [1]. Intermittency may also reflect the finding that poliovirus was isolated in most but not every stool sample collected from an identified, persistently infected individual [25]. (3) When enough surveillance sites are re-sampled for sufficiently long intervals it may even be possible to follow movements of the infected individual within large populations. This is illustrated by the shift from Jerusalem to Tel Aviv of the source of the second event (blue boxes in Table 1), and the shift within Tel Aviv of the source of the first event, i.e. the two times when progeny from the first event (pink boxes in Table 1) were isolated from the site on the Kav B branch, rather than along the Ayalon branch. And (4) The ability of environmental surveillance to determine the pattern of excretion within a region, specifically whether one individual or more than one is infected and when there is more than one, whether they live or at least excrete in different regions of the city. This is most clearly illustrated by the isolation of two unrelated VDPVs in the same sample (blue and pink box Table 1) from Reading site on March 14th2001 and the exclusive finding of isolates related to only one at simultaneously sampled upstream sites along the Ayalon trunk line. For reference, the Reading site is located downstream of the Ayalon and Kav B trunk lines from which most epidemiological event one and two VDPV2s were previously isolated, respectively.
Enteric viral surveillance
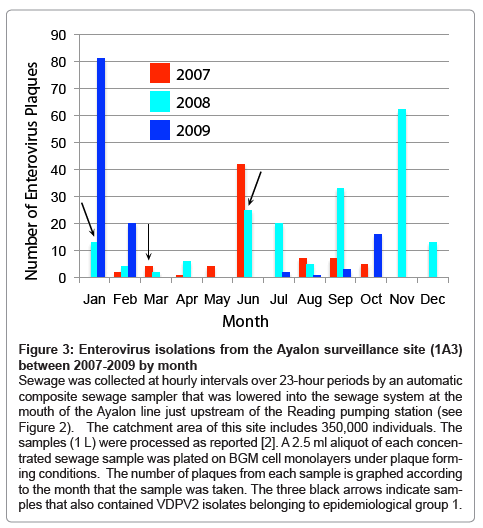
The WHO has recommended that a minimum of 30% of the sewage samples in a surveillance program be enterovirus positive to ensure that collection, transport and sample processing meet quality standards [11]. The annual rate of enterovirus positive samples in Israel has varied between 75 and 90% by plaque assay. Variations in the number of plaques provide a rough indication of the pattern of community enteroviral infections, since some enteroviruses may not grow on the cell line used for screening the sewage samples and others may not grow in tissue cultures at all. An example of the plaque counts from a single site representing a catchment population of 350,000 in central Israel over a 3-year period (2007 to 2009) is presented in Figure 3. Three samples also contained type 2 VDPVs (indicated by black arrows). Routine investigations of hospitalized cases from neurological wards for enteroviral infections compliment these findings and identify those viruses causing severe infections.
Figure 3: Enterovirus isolations from the Ayalon surveillance site (1A3) between 2007-2009 by month
Sewage was collected at hourly intervals over 23-hour periods by an automatic composite sewage sampler that was lowered into the sewage system at the mouth of the Ayalon line just upstream of the Reading pumping station (see Figure 2). The catchment area of this site includes 350,000 individuals. The samples (1 L) were processed as reported [2]. A 2.5 ml aliquot of each concen- trated sewage sample was plated on BGM cell monolayers under plaque form- ing conditions. The number of plaques from each sample is graphed according to the month that the sample was taken. The three black arrows indicate sam- ples that also contained VDPV2 isolates belonging to epidemiological group 1.
The viral etiology of acute gastroenteritis outbreaks between 2005 and 2011 has been systematically investigated, as has the etiology of all admissions between 2007 and 2011 of children less than five years of age for acute gastroenteritis at three sentinel hospitals in the northern Israel. These studies have provided information on the annual and seasonal pattern of severe rotavirus and norovirus infections in Israel [6,7].
Outbreak investigation requires close coordination between the National Center of Viral Gastroenteritis at the Central Virology Laboratory and District Health Officers and/or the staff of hospital Infectious Disease Departments. Real Time RT-PCR (rRT-PCR) was used to identify and quantify viral loads of norovirus infections, while rotavirus was identified by immuno-chromatographic (dipstick) assays and genotyped by RT-PCR. Stool suspensions from some of the outbreaks were first screened by electron microscopy (EM). Results based on size and morphology can be obtained in less than two hours using negative staining techniques and can focus the specific molecular identification tests that need to be carried out. On the down side, the limit of detection is 104 to 105 particles depending on the pathogen [26]. One hundred and one acute gastroenteritis outbreaks were investigated between 2005 and 2011. Genotype II.4 noroviruses accounted for all 67 (66.3%) of the outbreaks (Figure 4) and reovirus for another (1.0%). These outbreaks were due to the introduction of virus into groups of naïve individuals (Mother Nature). No viral etiology was found for 32 outbreaks (31.6%). Finally, stools from individual cases from one community-wide acute gastroenteritis outbreak contained different bacterial and viral agents including rotavirus and adenovirus and stools from some individuals actually contained more than on enteric pathogen. Subsequent investigation, in part directed by the pattern of pathogenic agents discovered during surveillance, revealed that bio- error (not bio-terror or Mother Nature) had caused the outbreak. In this case, inadvertent channeling of sewage into the community drinking water caused the outbreak.
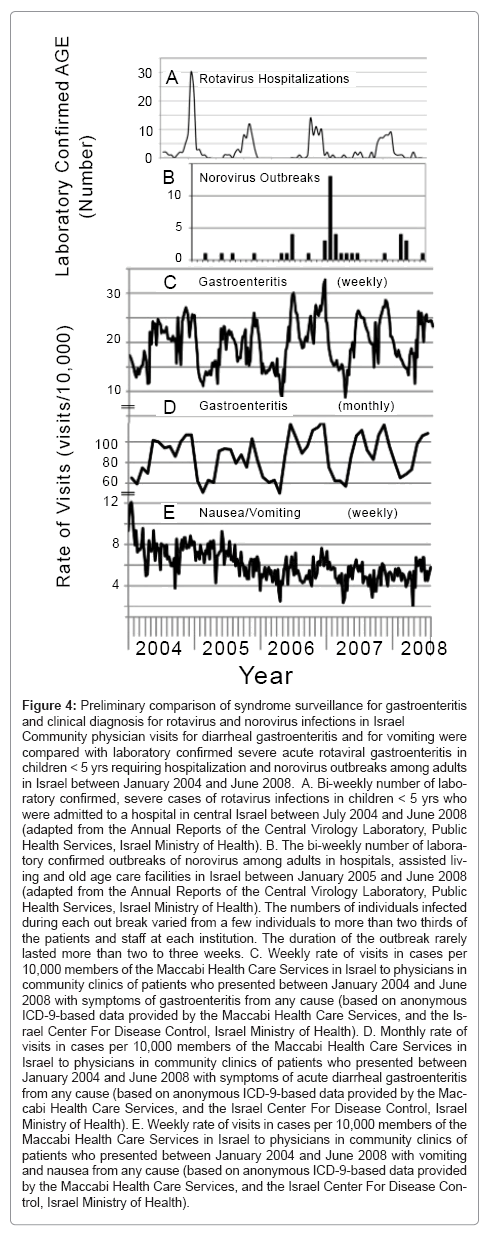
Figure 4: Preliminary comparison of syndrome surveillance for gastroenteritis and clinical diagnosis for rotavirus and norovirus infections in Israel
Community physician visits for diarrheal gastroenteritis and for vomiting were compared with laboratory confirmed severe acute rotaviral gastroenteritis in children < 5 yrs requiring hospitalization and norovirus outbreaks among adults in Israel between January 2004 and June 2008. A. Bi-weekly number of labo- ratory confirmed, severe cases of rotavirus infections in children < 5 yrs who were admitted to a hospital in central Israel between July 2004 and June 2008 (adapted from the Annual Reports of the Central Virology Laboratory, Public Health Services, Israel Ministry of Health). B. The bi-weekly number of labora- tory confirmed outbreaks of norovirus among adults in hospitals, assisted living and old age care facilities in Israel between January 2005 and June 2008 (adapted from the Annual Reports of the Central Virology Laboratory, Public Health Services, Israel Ministry of Health). The numbers of individuals infected during each out break varied from a few individuals to more than two thirds of the patients and staff at each institution. The duration of the outbreak rarely lasted more than two to three weeks. C. Weekly rate of visits in cases per 10,000 members of the Maccabi Health Care Services in Israel to physicians in community clinics of patients who presented between January 2004 and June 2008 with symptoms of gastroenteritis from any cause (based on anonymous ICD-9-based data provided by the Maccabi Health Care Services, and the Is- rael Center For Disease Control, Israel Ministry of Health). D. Monthly rate of visits in cases per 10,000 members of the Maccabi Health Care Services in Israel to physicians in community clinics of patients who presented between January 2004 and June 2008 with symptoms of acute diarrheal gastroenteritis from any cause (based on anonymous ICD-9-based data provided by the Mac- cabi Health Care Services, and the Israel Center For Disease Control, Israel Ministry of Health). E. Weekly rate of visits in cases per 10,000 members of the Maccabi Health Care Services in Israel to physicians in community clinics of patients who presented between January 2004 and June 2008 with vomiting and nausea from any cause (based on anonymous ICD-9-based data provided by the Maccabi Health Care Services, and the Israel Center For Disease Con- trol, Israel Ministry of Health).
The current protocols for concentration, amplification in tissue culture and analysis of polioviruses takes 14 to 21 days. Genotyping of individual enterovirus plaques from the sewage samples is labor and resource intensive and is not practical for rapid routine surveillance (see discussion of timeliness in relation to syndromic surveillance below). Studies are underway to test the possibility of drastically decreasing processing time by eliminating tissue culture amplification and trying different concentration strategies. For example, directly extracting the nucleic acids from concentrates prepared with current procedures that take less than two days will be compared to extraction from virus in ultracentrifugation pellets, prepared after mid-speed clarification to remove larger particulate matter. The advantage of eliminating the tissue culture amplification step is two fold, a large reduction in time and the fact that fastidious enteroviruses and other enteric viruses would be included in the population sequenced. The main disadvantages are a significant decrease in the amount of nucleic acid available for testing, mixtures of many closely related viruses and the need to make nucleic acid extractions from materials that are known to contain high amounts of inhibitors of RT and PCR enzymatic amplification reactions that might be co-extracted. The use of next generation sequencing that can be completed within days on some of the smaller platforms currently available, should overcome some of the difficulties from analyzing mixtures of viruses from sewage but would not be able to determine whether or not individual virus isolates were excreted by humans. Bar coding of samples would allow for simultaneous analysis of viruses from different samples, but at the cost of reduced depth of sequencing (the number of times that a given sequence is sequenced). The next generation sequencing techniques will include resequencing (specific primer based amplification in a multiplex reaction that simultaneously targeting many viral amplicons) and sequence independent deep sequencing (ligating amplification specific oligonucleotide sequences to the ends of randomly amplified and/or randomly sheared nucleic acids). Finkbeiner and colleagues demonstrated the feasibility of this approach by identifying known and previously unidentified enteric viruses in human stools collected from patients with acute gastroenteritis [27]. The challenge will be to succeed in sewage where viral concentrations will be orders of magnitude lower.
The screening of all admissions to the three sentinel pediatric departments of the three children’s hospitals to determine the etiology of severe acute gastroenteritis is part of an ongoing cooperative epidemiology study by the TAU-HCLV Study group (Tel Aviv University Epidemiology Department, the Hillel Yoffe, Carmel and Laniado Medical Centers, and the Central Virology Laboratory). The aim of the study was to characterize severe enteric infections in children before, during partial and after universal administration of rotavirus vaccines to newborn infants. This study has provided information on the economic burden caused by these viral agents [6] as well as preliminary information on rotavirus vaccine effectiveness [7]. It is mentioned here because the study has also provided annual molecular epidemiological surveillance data on the rotavirus and norovirus genotypes causing severe acute gastroenteritis in children and the prevalence and incidence of these viral infections that are of relevance for evaluating syndrome surveillance.
Syndrome surveillance
BT surveillance systems are usually based on analyzing changing patterns of symptoms and/or on programs based on the identification of specific pathogens [28]. Some examples of the latter have been described above. Their disadvantage is that they are limited to specific pathogens and would miss some bioterror or Mother Nature driven events. Syndrome surveillance has been studied as a means of identifying unusual or unexpected changes in rates of illness that may indicate the start of a bio-error, bio-terror or Mother Nature triggered event. Syndromic surveillance is predicated on (1) the observation that during these epidemiological events, people develop symptoms that lead to changes in behavior patterns (for example absence from work or school, increased purchases of over-the-counter palliative medicines), followed by visits to physicians where they present at first with general symptoms and later in the event with more specific symptoms, before the event is recognized and diagnosed [29] (2) the belief that it would be possible to develop effective methods to measure these changes in the pattern of symptoms as a means for early identification of the initiation of such an event and (3) that most pathogens that trigger bio-error, bio-terror or Mother Nature related events share symptoms with events caused by endemic or “routine” pathogens, particularly at early stages of their activity.
Two requirements for effective syndrome based surveillance are (1) timeliness of data acquisition, analysis, and reporting and (2) high sensitivity and specificity [28]. The potentially faster identification of unusual events through computer assisted syndromic surveillance compared with the labor intensive and time consuming surveillance for specific antigens described in the sections above and its ability to recognize unusual events because of symptom overlap with expected events are arguments in favor of its use or addition to existing surveillance programs. The number of people who will be infected and the difficulty in succeeding with intervention increase in relation to the time that elapses between initiation of the BT event and the notification that such an event is taking place [28].
A good case definition is needed to increase the signal to noise ratio [30]. However, too broad a definition will result in frequent false positive signals wasting limited resources and manpower in investigation. Conversely, too narrow a definition may exclude actual events. Integration of multiple data sources can significantly improve detection accuracy of syndrome-based surveillance systems [31]. Accuracy can be gauged and improved when syndromic surveillance programs are run in parallel with infectious agent surveillance. The use of laboratory confirmed influenza, especially pandemic influenza, to evaluate respiratory syndrome surveillance is one such example (see [28,32-34]). Some of these models have proven more successful at predicting Mother Nature triggered events than others. Similar evaluations can be conducted for acute gastroenteritis. Known pathogen driven events, such as laboratory confirmed hospital admissions with acute rotaviral gastroenteritis and norovirus outbreaks in adults, can be compared with community visits to physicians by patients presenting with acute diarrheal gastroenteritis or nausea or vomiting as shown for Israel (Figure 4). The most common cause for severe acute gastroenteritis in Israeli children under 5 years is rotavirus [6] followed by norovirus. The major symptom associated with rotavirus is diarrhea, while vomiting and nausea is more common for norovirus. There was a clear annual correlation between seasonality of severe rotaviral gastroenteritis requiring hospitalization in Israel (Figure 4a) and the second seasonal peak of physician visits (Figure 4C and D), In contrast there was none with the rate of visits for non-diarrheal vomiting and nausea. The causative agent of the first of the two annual diarrheal-triggered visits has not yet been identified. It is difficult to correlate norovirus outbreaks (Figure 4B) in adults with unusual activities of either diarrheal (Figure 4C and D) or non-diarrheal-triggered visits (Figure 4E) because of the low numbers of reported outbreaks. However, the small rise in visits at the end of the second peak of physician visits during the 2006-2007 winter season may reflect possible circulation of norovirus in the communities prior to the peak in a nationwide spike of norovirus outbreaks that occurred in hospitals and old age facilities between November 2006 and February 2007.
These preliminary comparisons for acute gastroenteritis in Israel illustrate some of the difficulties in validating syndromic surveillance. Monthly observations produce smoother curves (Figure 4C) than weekly observations (Figure 4D), but loose crucial timeliness required for bioterror surveillance. Lack of correlation may result from too restrictive a case definition (for example norovirus causes diarrhea in addition to vomiting and nausea). It may also arise when the categories compared are not completely equivalent. For example severe rotaviral gastroenteritis requiring hospitalization was compared with diarrheal-triggered community visits. Future validation should include clinical analysis of a representative sample of the less severe cases that are being monitored by the syndromic surveillance. As stated above. All Israelis are enrolled in one of four HMOs. All acute gastroenteritis visits to HMO physicians and to hospital emergency rooms are available for computerized prospective and retrospective syndrome surveillance. The information from the various enteric pathogen surveillance programs for enteric viral infections in Israel (described above) and initiation of laboratory surveillance of less severe community wide infections can and will be used to assess the quality of acute gastroenteritis based syndrome surveillance and help in fine-tuning the case definition.
One difficulty for evaluation of syndrome surveillance programs for detection of bioterror events is the lack of a reference standard for most currently envisioned infectious BT events [28]. However, symptoms during the initial stages of infections with many infectious BT agents resemble those at early stages of infections with natural pathogens. The more the resemblance, the more difficult it will be to recognize that the symptoms are associated with a BT event. On the other hand, the existence of “similar” natural infections suggests that they can serve as surrogate reference standards for BT events. Establishing the prevalence and incidence of usual illnesses over a number of years is required to establish the confidence limits of the seasonal pattern for a “normal” baseline for BT preparedness and for syndrome based surveillance programs [33].
Establishment of “normal” patterns can also serve as a yardstick for other public health programs. For example, the acute gastroenteritis study in the three sentinel hospitals in Israel and measurement of the rate of community wide physician visits for all causes of gastroenteritis (see above) established the baseline for measurement of the efficacy of the newly introduced rotavirus vaccination program [7] and will also serve as a baseline to determine whether the vaccination program exerts a selective pressure on the genotypes of the rotaviruses that still cause severe infections and on the relative incidences of severe rotavirus and norovirus infections.
This manuscript has focused on environmental and syndromic surveillance for enteric viral pathogens. Many potential pathogens that threaten mankind directly or indirectly by affecting domesticated fauna and flora either through bioerror, Mother Nature and especially bioterror can be transmitted through aerosols. Many pathogens are not even viral. As stated throughout this manuscript, the element of time is critical for identification and containment. Respiratory syndromic surveillance, mentioned above, is one important method that needs to be refined, expanded, and combined with pathogen detection and identification. In addition to more classical approaches discussed here for enteric pathogens, much effort has been devoted to the development of automatic semi-independent or autonomous remote rapid test systems that can provide warning in clinically relevant time frames. Included among such systems or surveillance programs that have already been tested and/or deployed are the Bio Watch (2003), Biohazard Detection (BDS), Biological Aerosol Sentry and Information System (BASIS; 2001-2), the Autonomous Pathogen Detection System (APDS; 2003) and Handheld Advanced Nucleic Acid Analyzer (HANAA) [https://www.globalseurity.org]. Monitoring using more common and widely dispersed items has included use of ventilation filters in public buildings [35] or airplanes [36]. Another area of active research is the development and testing of next generation sensor technology. These systems must take into account knowledge about the relation of particle size to transmission and detection [37] and the effect of environmental parameters on the survival of the pathogens [38] whether during transmission or during the interval between dispersal and analysis. This is especially true when a biological amplification step is required for detection
In conclusion, it is good to keep in mind as M. Stoto, Associate Director for Public Health at the RAND Center for Domestic and International Health Security pointed out, “An alarm system is only as good as what happens when the alarm goes off” [39].
Acknowledgements
We acknowledge the technical assistance (in alphabetical order) of B. Abramovitz, J. Alfandari, R. Azar. Ira Agabiev, T. Halmut, Y. Perepliotchikov, M. Hindiyeh, V. Levy and I. Silberstein. We would also like to acknowledge the members of the TAU-HCLV: Tel Aviv University (D. Cohen, Kh. Muhsen, G. Chodick, S. Goren, T. Ziv, A. Bialik, I. Pashai, W. Naamna, M. Brik), Hillel Yaffe Medical Center (E. Kasem, A. Kremer, F. Abu Rakia, E. Mahajna, M. Kaupstein, A. Athamna), Carmel Medical Center (M. Ephros, T. Zim, A. Kende), Laniado Medical Center (U. Rubinstein, J. Shachter, I. Weiss, A. Rimer, M. Israeli), and the Central Virology Lab (L. Shulman, I. Silberstein). The authors would like to thank the Israel Center for Disease Control and the Maccabi Health Care Services for providing anonymous data regarding outpatient visits to outpatient clinics for gastroenteritis
Funding
The Israel Ministry of Health funded most of the work described in this manuscript. The polio environmental surveillance involving simultaneous collections from multiple sites in central Israel in 2011 was part of grant 18-TSA- 032 for Post Eradication Sewage Surveillance - determining limits for detecting persistent polio excretors in large populations provided by the WHO, Geneva, Switzerland. The TAU-HCLV surveillance reviewed here was supported by grant V27-181-190 from the Department of Immunization, Vaccination, and Biologicals, WHO, Geneva, Switzerland.
References
- Hovi T, Shulman LM, van der Avoort H, Deshpande J, Roivainen M, et al. (2012) Role of environmental poliovirus surveilllance in global polio eradication and beyond. Epidemiol Infect 140: 1-13.
- Manor Y, Blomqvist S, Sofer D, Alfandari J, Halmut T, et al. (2007) Advanced environmental surveillance and molecular analyses indicate separate importations rather than endemic circulation of wild type 1 poliovirus in Gaza district in 2002. Appl Environ Microbiol 73: 5954-5958.
- Shulman LM, Manor Y, Handsher R, Delpeyroux F, McDonough MJ, et al. (2000) Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J Clin Microbiol 38: 3729-3734.
- Manor Y, Handsher R, Halmut T, Neuman M, Bobrov A, et al. (1999) Detection of poliovirus circulation by environmental surveillance in the absence of clinical cases in Israel and the Palestinian authority. J Clin Microbiol 37: 1670-1675.
- Shulman LM, Handsher R, Yang CF, Yang SJ, Manor J, et al. (2000) Resolution of the pathways of poliovirus type 1 transmission during an outbreak. J Clin Microbiol 38: 945-952.
- Muhsen K, Shulman L, Rubinstein U, Kasem E, Kremer A, et al. (2009) Incidence, characteristics, and economic burden of rotavirus gastroenteritis associated with hospitalization of israeli children <5 years of age, 2007-2008. J Infect Dis 200: S254-263.
- Muhsen K, Shulman L, Kasem E, Rubinstein U, Shachter J, et al. (2010) Effectiveness of rotavirus vaccines for prevention of rotavirus gastroenteritis-associated hospitalizations in Israel: a case-control study. Hum Vaccin 6: 450-454.
- Shulman LM, Silberstein I, Alfandari J, Mendelson E (2011) Genotyping rotavirus RNA from archived rotavirus-positive rapid test strips. Emerg Infect Dis 17: 44-48.
- Trujillo AA, McCaustland KA, Zheng DP, Hadley LA, Vaughn G, et al. (2006) Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J Clin Microbiol 44: 1405-1412.
- Hovi T, Shulman LM, H VDA, Deshpande J, Roivainen M, et al. (2012) Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol Infect 140: 1-13.
- WHO (2003) Guidelines for environmental surveillance of poliovirus circulation. WHO, Dept of Vaccines and Biologicals.
- Jorba J, Campagnoli R, De L, Kew O (2008) Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. J Virol 82: 4429-4440.
- Swartz TA (2008) The Epidemiology of Polio in Israel An Historical Perspective. Dyonon Pub. Ltd. 203 p.
- Slater PE, Orenstein WA, Morag A, Avni A, Handsher R, et al. (1990) Poliomyelitis outbreak in Israel in 1988: a report with two commentaries. Lancet 335: 1192-1195.
- WHO (1998) Scheme Adopted for Use for L20B Cells. Polio LaB Network Quarterly Update IV: 1-2.
- Wood DJ, Hull B (1999) L20B cells simplify culture of polioviruses from clinical samples. J Med Virol 58: 188-192.
- Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA (2005) Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol 59: 587-635.
- Shulman LM, Manor Y, Handsher R, Delpeyroux F, Halmut T, et al. (2001) Characterization of a highly evolved Sabin 2 polio strains isolated from sewage. Israel ISM News pp 34.
- Shulman LM, Manor Y, Sofer D, Handsher R, Swartz T, et al. (2006) Neurovirulent vaccine-derived polioviruses in sewage from highly immune populations. PLoS One 1: e69.
- Shulman LM, Sofer D, Manor Y, Mendelson E, Balanant J, et al. (2011) Antiviral activity of 3(2H)- and 6-chloro-3(2H)-isoflavenes against highly diverged, neurovirulent vaccine-derived, type2 poliovirus sewage isolates. PLoS One 6: e18360.
- Sutter RW, Kew OM, Cochi SL (2008) Poliovirus vaccine - live. In: Plotkin SA, Orenstein WA, Offit PA, editors. Vaccines, Fifth Edition. Philadelphia, Pa: Saunders/Elsevier. pp. 631-685.
- Kew OM, Wright PF, Agol VI, Delpeyroux F, Shimizu H, et al. (2004) Circulating vaccine-derived polioviruses: current state of knowledge. Bull World Health Organ 82: 16-23.
- Swartz TA, Green MS, Handscher R, Sofer D, Cohen-Dar M, et al. (2008) Intestinal immunity following a combined enhanced inactivated polio vaccine/oral polio vaccine programme in Israel. Vaccine 26: 1083-1090.
- Alexander JP Jr, Gary HE Jr, Pallansch MA (1997) Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J Infect Dis 175: S176-182.
- Kew OM, Sutter RW, Nottay BK, McDonough MJ, Prevots DR, et al. (1998) Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J Clin Microbiol 36: 2893-2899.
- Laue M, Bannert N (2010) Detection limit of negative staining electron microscopy for the diagnosis of bioterrorism-related micro-organisms. J Appl Microbiol 109: 1159-1168.
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, et al. (2008) Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 4: e1000011.
- Bravata DM, McDonald KM, Smith WM, Rydzak C, Szeto H, et al. (2004) Systematic review: surveillance systems for early detection of bioterrorism-related diseases. Ann Intern Med 140: 910-922.
- Stoto MA (2005) Syndromic surveillance. Issues in Science and Technology 21: 49-56.
- Marsden-Haug N, Foster VB, Gould PL, Elbert E, Wang H, et al. (2007) Code-based syndromic surveillance for influenzalike illness by International Classification of Diseases, Ninth Revision. Emerg Infect Dis 13: 207-216.
- Wang L, Ramoni MF, Mandl KD, Sebastiani P (2005) Factors affecting automated syndromic surveillance. Artif Intell Med 34: 269-278.
- Wallace DJ, Arquilla B, Heffernan R, Kramer M, Anderson T, et al. (2009) A test of syndromic surveillance using a severe acute respiratory syndrome model. Am J Emerg Med 27: 419-423.
- Kaufman Z, Aharonowitz G, Dichtiar R, Green MS (2006) Estimating the usual prevalence and incidence of acute illness in the community: implications for pandemic influenza and bioterrorism preparedness. Isr Med Assoc J 8: 563-567.
- Moore K, Black J, Rowe S, Franklin L (2011) Syndromic surveillance for influenza in two hospital emergency departments. Relationships between ICD-10 codes and notified cases, before and during a pandemic. BMC Public Health 11: 338.
- Goyal SM, Anantharaman S, Ramakrishnan MA, Sajja S, Kim SW, et al. (2011) Detection of viruses in used ventilation filters from two large public buildings. Am J Infect Control 39: e30-38.
- Korves TM, Johnson D, Jones BW, Watson J, Wolk DM, et al. (2011) Detection of respiratory viruses on air filters from aircraft. Lett Appl Microbiol 53: 306-312.
- Gralton J, Tovey E, McLaws ML, Rawlinson WD (2011) The role of particle size in aerosolised pathogen transmission: a review. J Infect 62: 1-13.
- Tang JW (2009) The effect of environmental parameters on the survival of airborne infectious agents. J R Soc Interface 6: S737-746.
- Eban K (2007) Biosense or Biononsense? The Scientist 21: 33-38.
Relevant Topics
- Anthrax Bioterrorism
- Bio surveilliance
- Biodefense
- Biohazards
- Biological Preparedness
- Biological Warfare
- Biological weapons
- Biorisk
- Bioterrorism
- Bioterrorism Agents
- Biothreat Agents
- Disease surveillance
- Emerging infectious disease
- Epidemiology of Breast Cancer
- Information Security
- Mass Prophylaxis
- Nuclear Terrorism
- Probabilistic risk assessment
- United States biological defense program
- Vaccines
Recommended Journals
Article Tools
Article Usage
- Total views: 14749
- [From(publication date):
specialissue-2012 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 10172
- PDF downloads : 4577