Research Article Open Access
Biodegradation of Polyethylene by a Soil Bacterium and AlkB Cloned Recombinant Cell
Moon Gyung Yoon, Hyun Jeong Jeon and Mal Nam Kim*Department of Life Science, Sangmyung University, Seoul 110-743, Korea
- *Corresponding Author:
- M. N. Kim
Department of Life Science, Sangmyung University
Seoul 110-743, Korea
Tel: +82-2-2287-5150
Fax: +82-2-2287-0070
E-mail: mnkim@smu.ac.kr
Received date: March 10, 2012; Accepted date: April 13, 2012; Published date: April 15, 2012
Citation: Yoon MG, Jeon HJ, Kim MN (2012) Biodegradation of Polyethylene by a Soil Bacterium and AlkB Cloned Recombinant Cell. J Bioremed Biodegrad 3:145. doi: 10.4172/2155-6199.1000145
Copyright: © 2012 Yoon MG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
A mesophilic bacterium capable of Low-Molecular-Weight Polyethylene (LMWPE) biodegradation was isolated from a beach soil having been contaminated extensively with crude oil. The isolated strain was rod-shaped gram negative bacterium and was identified as Pseudomonas sp. E4 through the 16S rDNA sequencing. The biodegradability test in the compost inoculated with the isolated strain for LMWPE having weight-average-molecularweight (Mw) in the range of 1,700~23,700 indicated that 4.9~28.6 % of the carbon was mineralized into CO2 after 80 days at 37°C. The biodegradability decreased with increase in Mw of LMWPE. In comparison to other previous works which reported biodegradation of pre-oxidized polyethylene, we investigated biodegradation of non-oxidized LMWPE whose Mw was well above the Mw upper limit penetrable through microbial membrane. The smooth surface of LMWPE sheet became eroded as a result of the biodegradation, and the degree of the surface erosion became more pronounced for LMWPE with lower molecular weight in line with the biodegradability test results. The alkane hydroxylase gene (alkB) of Pseudomonas sp. E4 was expressed in Escherichia coli BL21 and the recombinant E. coli BL21 mineralized 19.3% of the carbon of LMWPE into CO2 during the biodegradation in the compost at 37°C for 80 days, while the recipient cell was not active at all toward the LMWPE biodegradation, indicating that the alkB from Pseudomonas sp. E4 plays a central role in the LMWPE degradation even in the absence of the other specific enzymes like rubredoxin and rubredoxin reductase.
Keywords
Polyethylene; Biodegradation; Soil bacterium; Isolation; alkB; Gene cloning
Introduction
Worldwide production of plastics is increasing by approximately 5% every year [1], because plastics exhibit advantages such as clarity, hardness, processability and lightness together with price competiveness. However plastics are mostly made from fossil resources such as petroleum, coal and natural gas [2] and lots of plastics are discarded after short service life [3]. This aroused serious public concern so that that mankind classified plastics as one of the worst inventions ever made.
Waste plastics lay enormous burden on the environment, because their recalcitrance to degradation accelerates the accumulation in nature. Waste plastics buried in soil cause the water clogging phenomena and devastate soil for agricultural cultivation. Lots of animals die of waste plastics either by being caught in the waste plastics trap or by swallowing the waste plastics debris to exert ruinous effects on the ecosystem [4].
Recycling of waste plastics can be a good alternative solution. However, collection and cleansing of waste plastics are an expensive process, and the physical properties decay seriously during the reprocessing procedure [5].
Polyethylene is highly hydrophobic and chemically inert, and microbes on the earth surface have not yet been fully evolved to digest the artificially made plastics. A lot of research has been carried out to alleviate the environmental burden by improving degradability of the waste polyethylene. Abiotic pretreatment such as weathering, UV irradiation and thermal treatment was employed to raise the hydrophilicity of polyethylene by introducing polar groups such as carbonyl groups to the polyethylene backbone chain and thus facilitates the microbes to metabolize the unwieldy plastics [6,7]. A few microbes capable of degradation of the pretreated polyethylene have so far been isolated from soil, sea water, compost and activated sludge. These include Penicillium simplicissimum, Brevibacillus borstelensis, Rhodococcus rubber, Rh. rhodochrous, Rh. erythropolis, Bacillus circulans, B. brevies, B. sphaericus, B. pumilus, B. halodenitrificans, B. cereus, Delftia acidovorans, Flavobacterium sp., Staphylococcus epidermidis, Arthrobacter koreensis, A. paraffineus, Pseudomonas aeruginosa, Shewanella putrefaciens, Ralstonia basilensis and Nocardia asteroids [3,8-21].
Microbes biodegrading pristine polyethylene have never been isolated so far, and thereby we employed an alkane, i.e., hexadecane, whose basic chemical structure is identical to that of polyethylene, as a model compound for the investigation of the polyethylene biodegradation and the relevant genes.
Various enzymes are involved in the hexadecane metabolic pathway of G. thermodentificans [22]. Hexadecane metabolic pathway flows into the TCA cycle after the hexadecane degradation by the relevant degrading enzyme through the β-oxidation [23].
Belhaj et al. [24] studied alkB and alkB related genes encoding an alkane degrading enzyme, alkane hydroxylase, in P. aeruginosa isolated from a soil contaminated with crude oil. Saadoun et al. [25] identified alkB in Streptomyces exhibiting a high activity toward alkane degradation. Feng et al. [22] isolated a thermophilic strain, Geobacillus thermodenitrificans NG80-2, from a soil around oil storage tanks. The strain was highly active for hexadecane degradation. They confirmed alkane hydroxylase protein expression by proteomics, and suggested the strain as an eligible tool for the bioremediation of oil contaminated soil. Piccolo et al. [26] described the alkane degradation enzyme system of Gordonia sp. which can grow on C12~ C36 n-alkanes.
Reddy et al. [27] revealed that the β-carbons of hexadecane were oxidized repetitively into carboxylic acid groups and the alkane chain was cleaved by way of the formation of the acetyl-coenzyme A.
In comparison to the well investigated alkB, encoding the alkane hydroxylase which catalyzes the first step of the alkane degradation process, few studies have been carried out for the other degradation genes [23,28-30]. Marchant et al. [31] reported that the hexadecane degradation encoding alkB of Geobacillus thermoleovorans T70, isolated from the Northern Ireland forest soil, and exhibited 96% of homology in comparison to that of R. erythropolis. Smits et al. [32] detected the alkB by using the alkB primer from the 18 strains such as Pseudomonas oleovorans, P. putida, P. fluorescens, P. aeruginosa, Acinetobacter spp., A. calcoaceticus, Comamonas testosteroni, Stenotrophomonas maltophilia, Ralstonia eutropha, R. rhodochrous , R. erythropolis and Amycolatopsis rugosa. The DNA sequence alignment and phylogenetic analyses revealed that the DNA sequence identity was in the range between 36.0 and 99.8% depending on the extent of difference in the microbial genera.
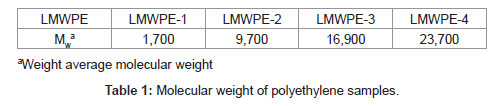
In this study, isolation of a strain capable of the polyethylene degradation was attempted from a crude oil contaminated soil at room temperature. Instead of pre-oxidized polyethylene which has been used for the isolation of polyethylene-degrading strains in the previous studies, we used LMWPE which had been prepared by thermal degradation of a commercially available neat polyethylene under a strict nitrogen atmosphere. The weight average molecular weight (Mw) of LMWPE was in the range between 1,700 and 23,700 as shown in Table 1, which was far superior to 500, the maximum value reported to be penetrable through microbial membrane. Biodegradability of LMWPE with different Mw was assessed in the compost inoculated with the isolated strain after sterilization. In comparison to the previous studies which employed n-alkanes, whose molecular weight was less than 500, to investigate the genes involved in degradation of n-alkanes [26], here, the alkB from the isolated strain was expressed in E. coli BL21 to see whether the host cell acquires the activity to mineralize LMWPE whose molecular weight is well over 500.
Materials and Methods
Isolation of the polyethylene degrading microorganisms
Isolation of the LMWPE degrading strains was performed according to the method proposed by Hadad et al. [12] after a condign modification. The enrichment medium for the isolation of the LMWPE degrading strains was composed of KNO3 2.0 g, K2HPO4 3.7 g, KH2PO4 5.8 g, MgSO4∙H2O 0.25 g, yeast extract 0.1 g, with l L of distilled water. The final pH was adjusted to 7.8. Ultra-sonication was performed to obtain the well dispersed medium. The soil samples were collected from Manripo-beach, Yellow sea, Korea, where a serious oil spill accident had taken place. The soil sample suspension was inoculated into the enrichment medium. LMWPE with Mw of 1,700 at a concentration of 1 mg/ml was added to the medium, which was then incubated at 37°C for 7 days in a rotary shaker. Several times of the subcultures were made by inoculating the original culture to the fresh medium containing LMWPE as a sole source of carbon.
After the subcultures, the enriched culture broth was spread on the agar plates supplemented with 1 ml/l of emulsified hexadecane and incubated at 37°C for 1 week and then the size of the clear zone formed by the strain was measured. The emulsified agar plate for the selection of the polyethylene degrading strains was prepared by adding 1 ml of hexadecane (Sigma) into 1L of the enrichment medium with 0.1 g of a detergent (Plysurf A210G). After emulsification of the medium by using an ultra-sonicator (280 W, 90 min), agar (15 g/L) was added to the medium [33].
Hexadecane was used instead of LMWPE for the preparation of the agar plates, because the clear zone formed was far less clearly seen when LMWPE was dispersed in the agar plate instead of hexadecane.
LMWPE-degrading microorganisms were selected based on the size of the clear zone.
Morphology of the isolated strain was investigated by using a Scanning Electron Microscope (SEM; JEOL, Tokyo, Japan, Model JSM- 5600LV).
Phylogenetic identification of the polyethylene degrading bacterium
The isolated bacterium was identified by using the 16S rDNA sequencing. The universal primer used was: forward 518F (5’-CCAGCAGCCGCGGTAATACG-3’) and reverse 800R (5’-TACCAGGGTATCTAATCC-3’) [34]. Amplified nucleotide sequences were determined with National Center for Biotechnology Information (NCBI) BLAST database (http://www.nih.nov.ncbi). The sequences were aligned together with those of the representative members of the selected genera with the CLUSTAL W program [35]. The phylogenetic tree was inferred from the neighbor-joining method with MEGA version 3.1 [36].
Detection of the alkB
The Polymerase Chain Reaction (PCR) was performed to amplify the alkB of the isolated strain; 35 cycles of pre-denaturation at 94°C for 5 min, denaturation at 94°C for 1 min, annealing at 40°C for 30 s, and extension at 72°C for 30 s, finally extension at 72°C for 7 min. The PCR products were identified by performing the gel electrophoresis on the 1.5 % agarose (SeaKem; 100 Vx30 min).
Cloning of the alkB
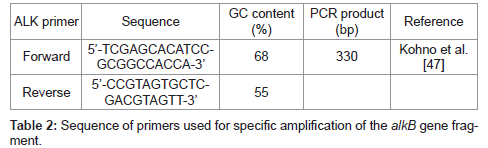
The alkB was amplified using the primer having the restriction enzyme site of EcoRI (G ← AATTC) and HindIII (A ← AGCTT) (Enzynomics, Korea). The amplified PCR products were purified by using the QIAquick gel extraction Kit (Qiagen), and the pUC19 was extracted by using the plasmid purification Kit (Intron) [37]. The plasmid vector and the PCR products were treated with the two restriction enzymes, EcoRI and Hind III, at 37°C, and ligated by using the ligase. The ligated recombinant plasmid was transformed to E. coli BL21, and then the white colony through the blue/white screening was chosen as the alkB cloned cell [38]. Using the white colony, the PCR was performed to amplify the alkB using the primer in Table 2, and the analysis of the amplified gene by using NCBI BLAST database proved the white colony to be the alkB cloned recombinant cell.
Biodegradation of LMWPE by the isolated bacterium and the alkB cloned recombinant cell
The isolated bacterium incubated in Luria-Bertani Broth at 37°C for 24 h was inoculated to the previously sterilized compost [33]. The rate of CO2 production was measured according to KS M3100-1:2002; MOD ISO 14855:1999 [39] at 37°C for 80 days. The activity of the alkB cloned recombinant cell toward LMWPE degradation was assessed following the same method used for the activity measurement of the isolated strain.
Detection of the viable alkB cloned recombinant cell
In order to confirm the survival of the alkB cloned recombinant cell, 10 g of the compost harvested after 80 days of the biodegradation test was suspended in 1 L of demineralized water. The supernatant was inoculated to Luria-Bertani (LB) agar medium (Difco) containing ampicillin. After the incubation, the white colony was selected through the blue/white screening. Its alkB was amplified by the PCR using the primer in Table 2. The amplified gene was analyzed by using the NCBI BLAST database.
Analyses of the alkB sequence
The alkB cloning gene nucleotide of the isolated bacterium was analyzed by using the NCBI BLAST database. The alkB homology was examined by aligning the gene sequence using the Seq men and Megalign based CLUSTAL W program [40] and the T-Coffee program [41].
Results and Discussion
Isolation of the LMWPE degrading microorganism and the phylogenetic identification
The mesophilic bacterium being active for the LMWPE biodegradation was isolated from a soil at a Yellow Sea beach of the Korea peninsula where the serious crude oil contamination had taken place [42]. The isolated strain was rod-shaped gram negative bacterium with 1.2× 0.5 μm in size.
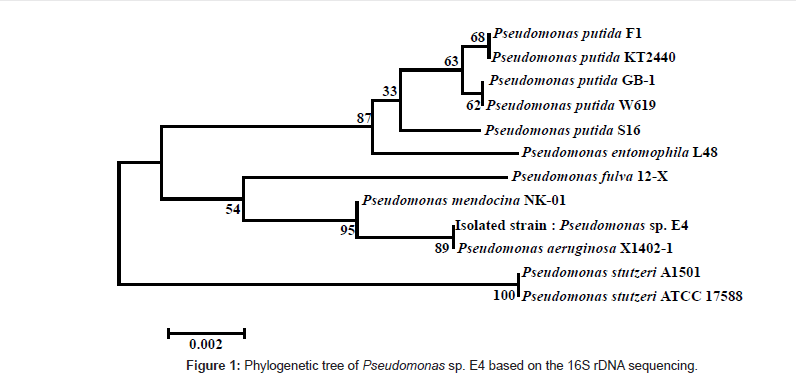
The isolated strain was identified through the 16S rDNA sequencing, and 1356 out of 1400 bp coincided with those of Pseudomonas sp. Figure 1 shows the phylogenetic tree for the isolated strain by using the BLAST search tool of NCBI web server, based on the analyses of the 16S rDNA sequence, and on the homology between the 16S rDNA sequences of the registered strains.
Chaerun et al. [43] isolated crude oil-utilizing bacteria from 3 beaches of Japan where the oil spill accident occurred in 1997 by the Russian oil tanker, Nakhodka. Most of their isolated strains belonged to P. aeruginosa with 99% of homology. They grew well in the medium containing crude oil as a sole carbon source. The isolated strains utilized long-chain alkanes effectively but not aromatic compounds. G. thermodenitrificans NG80-2 isolated by Feng et al. [22] from the soil near an oil storehouse was also able to degrade long-chain alkanes efficiently.
In comparison to n-alkanes, polyethylene has been deliberately oxidized by compounding with photo-oxidation catalyst followed by exposure to UV or sunlight or at least compounding with biodegradable polymers such as Bionolle® and starch was performed for the microbial analyses [44]. Yamada-Onodera et al. [9] and Hadad et al. [12] ferreted out P. simplicissimum YK and B. borstelensis, respectively, by cultivating at 50°C for 2 weeks on the agar plate containing preoxidized polyethylene as a sole carbon source, from the soil where polyethylene waste had been buried. Orr et al. [11] identified R. rubber, an aerobic, non-sporulating gram positive bacterium, as a polyethylene degrading bacterium.
LMWPE biodegradation activity of the isolated bacterium
LMWPE prepared by thermal decomposition of High Density Polyethylene (HDPE) and Low Density Polyethylene (LDPE) under inert atmosphere was subjected to the biodegradability test in the compost at 37°C under the controlled conditions according to KS M3100-1:2002; MOD ISO 14855:1999. Since the chemical reaction, i.e., microbial transformation of polyethylene, took place extremely slowly, the chemical reaction enthalpy should not lay a significant temperature gradient through the biodegradability test vessel, if any, so that the inside temperature of the test vessel is almost the same as the outside one.
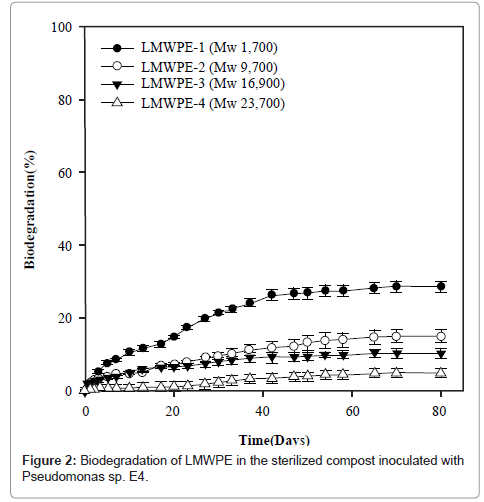
Figure 2 demonstrates the biodegradability of LMWPE in the compost at 37°C. Here the biodegradability was defined as follows:
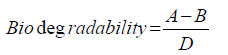
Where A is the accumulated amount of CO2 evolved from the compost loaded with LMWPE, and B is the accumulated amount of CO2 evolved from the compost without LMWPE. D, 3.14 g of CO2/g of PE, is the amount of CO2 evolved when all the carbons in the PE samples were assumed to be mineralized into CO2 . The compost contained 5 wt% of the polyethylene on dry basis. It had been sterilized prior to the inoculation with Pseudomonas sp. E4. LMWPE had weight average molecular weight (Mw) in the range of 1,700~23,700. PE-1, PE-2, PE-3 and PE-4 showed 28.6%, 14.9%, 10.3% and 4.9% of biodegradability, respectively, after 80 days of the biodegradation at 37°C in the compost, indicating that the biodegradability decreased with increase in the molecular weight of LMWPE.
LMWPE began to degrade slowly without any induction period. However, the LMWPE biodegradation ability of the isolated strain was lower than that of Chelatococcus sp. E1 who degraded 44.5% of PE-1 during the same period of time as reported in our previous study (data not shown). This was attributed in part to the fact that the biodegradation of PE-1 by Chelatococcus sp. E1 was carried out at 58°C. Abiotic degradation of the LMWPE should occur concomitantly with the biodegradation in the compost and the abiotic degradation should occur to a larger extent at 58°C than at 37°C. The abiotically degraded LMWPE should be more susceptible to microbial attack than the intact LMWPE.
Decrease in the biodegradability with increase in the molecular weight has also been observed in many other polymers including biodegradable polymers. Teeraphatpornchai et al. [45] employed Paenibacillus amylolyticus for the biodegradation of PLA having Mw in the range of 5,000~20,000. The strain formed smaller clear zone as the molecular weight of PLA increased. Koutny et al. [46] reduced the molecular weight of HDPE through photo- and thermo-degradation. Besides the reduction in the molecular weight, lots of carboxylic groups were formed as a result of the abiotic degradation. And the resulting low-molecular-weight HDPE was digested more easily by R. rhodochrous and N. asteroides.
Morphology of the LMWPE sheets after the biodegradation
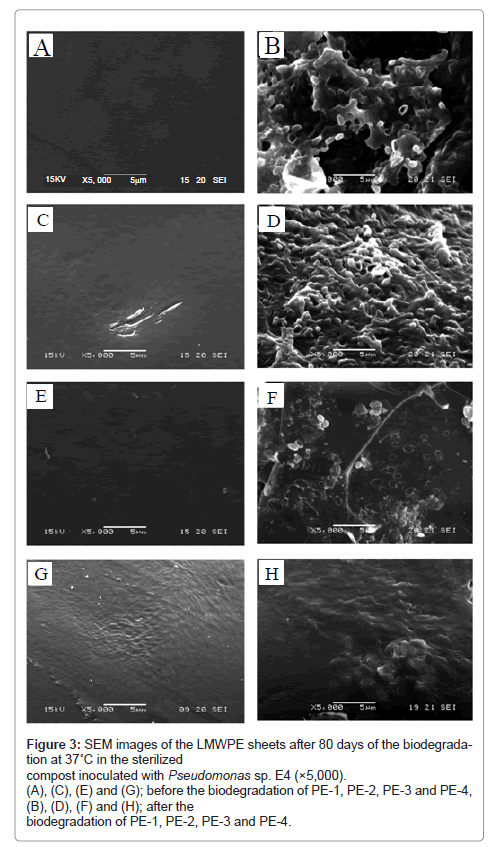
Morphology of LMWPE after the biodegradation in the compost inoculated with the isolated bacterium for 80 days at 37°C was examined by using SEM as shown in Figure 3. The smooth surface of the LMWPE sheets became eroded as a result of the biodegradation. A lot of the bacterial cells adhering to the surface of the LMWPE sheets can be seen. In line with the biodegradability results in Figure 2, the degree of the surface erosion became more pronounced for LMWPE with lower Mw.
Figure 3: SEM images of the LMWPE sheets after 80 days of the biodegradation at 37˚C in the sterilized compost inoculated with Pseudomonas sp. E4 (×5,000). (A), (C), (E) and (G); before the biodegradation of PE-1, PE-2, PE-3 and PE-4, (B), (D), (F) and (H); after the biodegradation of PE-1, PE-2, PE-3 and PE-4.
Bonhomme et al. [10] also demonstrated that R. rhodochrous and N. asteroides GK911 eroded polyethylene sheet and grew on the whole wall made by the strains. The polyethylene sheet was exposed to the sunlight at 60°C for 300 h prior to the cultivation with the strains for 2 months.
Analyses and genetic homology of the alkB sequence
To our best knowledge, microbial enzyme genes for polyethylene have never been reported. Instead, alkane degradation enzymes have been studied a lot. Since gene encoding alkane hydroxylase is key enzymes catalyzing the first step reaction of the alkane degradation [45], the same kind of the enzyme may participate also in the polyethylene degradation.
We tried to amplify the alkane hydroxylase gene by using the PCR primer which had been employed for the detection of Pseudomonas and Rhodococcus spp. belonging to Group III classified by Kohno et al. [47].
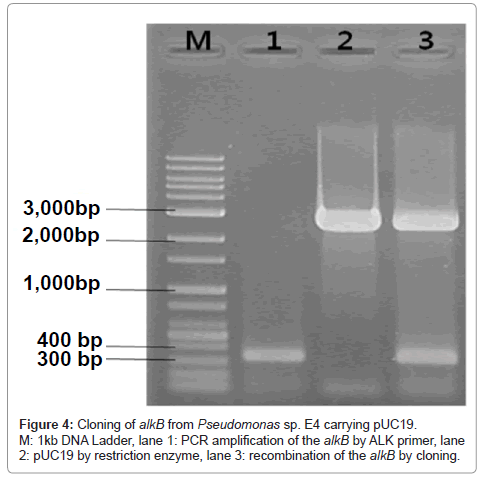
Amplified alkB segments were subjected to the electrophoresis on the agarose gel and a band appeared at 330 bp as demonstrated in Figure 4 lane 1.
Belhaj et al. [24] isolated P. aeruginosa from the soil which had been contaminated with hydrocarbons. They examined the growth rate of the strain in the medium composed of low molecular weight alkanes such as hexane, heptane, octane and decane. Investigation of the alkB cloned strain inserted pCR2.1 vector and sequence tags of the alkB genes revealed that the alkB was extremely diverse.
Kloos et al. [48] performed the PCR for the total DNA of the soil sample at Bavaria, Germany. They obtained 58 PCR products whose bands appeared at 550 ± 50 bp. The hybridization of the PCR products yielded 42 alkB clones attached with the alkB complementary probe. Investigation of the alkB homology using the cDNA library cloning revealed that the genetic homogeny between the 42 alkB clones was in the range of 57~90 %. On the other hand, the PCR for the total DNA of the agricultural soil sample detected 21 alkB genes among which the 2 alkB genes showed genetic difference less than 1% and the 19 alkB genes exhibited the genetic homogeny in the range of 50~90%.
Heiss-Blanquet et al. [49] measured the amount of the gene in the soil samples to assess the ecological importance of the bacteria for the degradation of alkanes of short-chain length by using the alkB. They observed that the alkB depended on the environmental and ecological state and that the amount of the alkB was larger in the soil contaminated with hydrocarbons. Almost all the genes identified as alkB existed in P. aeruginosa, P. putida and P. mendocina.
Saadoun et al. [25] isolated microorganisms from an oil polluted soil and investigated their growth using diesel oil as a sole carbon source. They confirmed the presence of the alkB through the PCR analyses. Two groups of the bands appeared at 316~334 bp and 460~550 bp, respectively, on the agarose gel, and the alkB genes exhibited bands between 320 and 550 bp. Smits et al. [32] observed that a single alkBspecific primer cannot detect all the alkane degrading microorganisms due to the microbial species-specificity in the detection of alkane degraders through the PCR technique. The alkB was found to be totally different depending on the microbial species due to the large difference in the alkB sequence. For this reason, Kohno et al. [47] prepared 3 different PCR primers and amplified them to apply for the detection of the alkane hydroxylase gene in the exploration of the alkane degrading microorganisms. They classified the detected microorganisms into 3 groups: Group I with 185 bp was comprised of Acinetobacter, Pseudomonas and Corynebacterium spp. belonged to Group II with 271 bp and Group III with 330 bp covered Pseudomonas and Rhodococcus spp.
Lo Piccolo et al. [26] discussed that the alkane molecule should slide into the microbial hydrophobic pocket to have the terminal methyl group be hydroxylated to initiate the biotransformation of the alkane molecule.
Since the band of the amplified alkB segments of the present isolated strain appeared at 330 bp as shown in Figure 4, in accordance with the Group III PCR products obtained by Kohno et al. [47], it can be said that the present strain belongs to Pseudomonas sp.
Comparison of the alkB of Pseudomonas sp. E4 with those of P. aerugionosa PA7 (NC009656.1), P. aerugionosa PAO1 (NC002516.2) and P. mendocina (NC009439.1) which are registered in NCBI yielded about 86% of homology as shown in Figure 5.
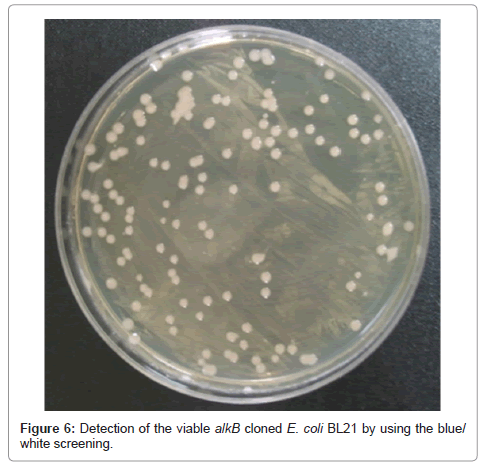
Detection of viable alkB cloned recombinant cell
The viability of the alkB cloned recombinant cell was examined after the LMWPE biodegradation. Figure 6 demonstrates that the cells survived the 1st screening in the LB plate containing ampicillin, and that the white colonies were detected after the 2nd blue/white screening. This clearly confirms that the alkB cloned recombinant cell remained viable. The PCR products of the strain appeared at 330 bp, and comparison with the NCBI BLAST database revealed that the strain possessed the alkB. Therefore it can be said that the alkB cloned recombinant cell survived the biodegradation test for 80 days.
LMWPE biodegradation activity of the alkB cloned recombinant cell
Piccolo et al. [26] observed that a Gram positive bacterium, Gordonia sp., was able to grow on n-alkanes from C12 to C36. The recombinant Streptomyces coelicolor M145 expressing the Gordonia alkB gene grew on n-triacontane (C36) as the sole carbon source [50].
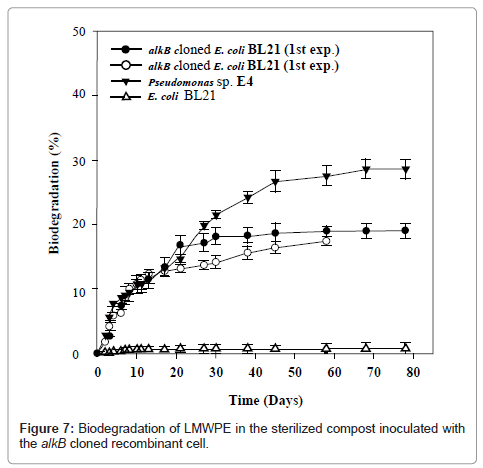
In order to see whether the alkB from Pseudomonas sp. E4 is also active for the biotransformation of LMWPE whose molecular weight is well over that of triacontane, the activity of the alkB cloned recombinant cell was examined toward the LMWPE biodegradation in the compost under controlled conditions as shown in Figure 7. It is to be noted here that the recipient cell, E. coli BL21, was not active at all toward the LMWPE biodegradation. In contrast, the alkB cloned recombinant cell mineralized 19.3% of the carbon of PE-1 into CO2 at 37°C for 80 days. A duplicated experiment yielded almost the same result. The accumulated amount of CO2 produced by Pseudomonas sp. E4 was almost the same as that produced by the alkB cloned recombinant cell up to 20 days. Therefore it can be said that alkB was well cloned to E. coli BL21 and that the host cell acquired the activity for the mineralization of LMWPE.
Conclusions
A mesophilic rod-shaped gram negative bacterium active for the LMWPE biodegradation was isolated from a seashore soil sample and was identified as Pseudomonas sp. through the 16S rDNA sequencing. The isolated strain mineralized LMWPE whose Mw was in the range of 1,700~23,700 with appreciable activity at 37˚C in the compost under the controlled conditions. However, the biodegradation activity decreased with increase in the molecular weight of LMWPE. Since microbial enzyme genes for the polyethylene degradation have never been reported, we employed an n-alkane degrading enzyme for the investigation of LMWPE degrading enzyme gene using hexadecane as a model compound instead of LMWPE. The amplified alkB segments showed a band at 330 bp on the agarose gel and coincided with the Group III PCR products as classified by Kohno et al. [47]. The alkB cloned recombinant cell was viable even after the biodegradation tests at 37°C for 80 days. The alkB cloned recombinant cell was as active as Pseudomonas sp. E4 toward the LMWPE biodegradation in the early stage of the biodegradation, while the recipient cell, E. coli BL21, was not active at all, indicating that the alkB from Pseudomonas sp. E4 played a central role in the mineralization of LMWPE into CO2.
Acknowledgement
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-A001-0021).
References
- Mani M, Subash C, Nagarajan G (2009) Performance, emission and combustion characteristics of a DI diesel engine using waste plastic oil. Appl Thermal Eng 29: 2738-2744.
- Seymour RB (1989) Polymer science before & after 1899: Notabledevelopments during the lifetime of Maurtis Dekker. Journal of Macromolecular Science: Part A - Chemistry 26: 1023-1032.
- Sudhakar M, Doble M, Murthy PS, Venkatesan R (2008) Marine microbe-mediated biodegradation of low-and high-density polyethylenes. Int Biodeterior Biodegradation 61: 203- 213.
- Conner DK, O’dell R (1998) The tightening net of marine plastics pollution. Environment: Science and policy for sustainable development 30: 16-36.
- Zhao JH, Wang XQ, Zeng J, Yang G, Shin F, et al. (2005) Biodegradation of poly(butylene succinate-co-butylene adipate) by Aspergillus versicolor. Polym Degrad Stab 90: 173-179.
- Albertsson AC, Andersson SO, Karlsson S (1987) The mechanism of biodegradation of polyethylene. Polym Degrad Stab 18: 73-87.
- Gröning M, Hakkarainen M, Albertsson AC (2004) Recycling of glass fibre reinforced phenolic prepreg waste. Part 1: Recovery and reuse of glass fibres in PP and PA6. Polym Polym Compos 12: 491-500.
- Albertsson AC, Erlandsson B, Hakkarainen M, Karlsson S (1998) Molecular weight change and polymeric matrix change correlated with the formation of degradation products in biodegraded polyethylene. J Environ Polym Degrad 6: 187-195.
- Yamada-Onodera K, Mukumoto H, Katsuyaya Y, Saiganji A, Tani Y (2001) Degradation of polyethylene by a fungus, Penicillium simplicissimum YK. Polym Degrad Stab 72: 323-327.
- Bonhomme S, Cuer A, Delort AM, Lemaire J, Sancelme M, et al. (2003) Environmental biodegradation of polyethylene. Polym Degrad Stab 81: 441- 452.
- Orr IG, Hadar Y, Sivan A (2004) Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber. Appl Microbiol Biotechnol 65: 97-104.
- Hadad D, Geresh S, Sivan A (2005) Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J Appl Microbiol 98: 1093- 1100.
- Sivan A, Szanto M, Pavlov V (2006) Biofilm development of the polyethylene-degrading bacterium Rhodococcus ruber. Appl Microbiol Biotechnol 72: 346- 352.
- Marchal R, Nicolau E, Ballaguet JP, Bertoncini F (2008) Biodegradability of polyethylene glycol 400 by complex microfloras. Int Biodeterior Biodegradation 62: 384-390.
- Reddy MM, Gupta RK, Gupta RK, Bhattacharya SN, Parthasarathy R (2008) Abiotic oxidation studies of oxo-biodegradable polyethylene. J Polym Environ 16: 27-34.
- Roy PK, Titus S, Surekha P, Tulsi E, Deshmukh C, et al. (2008) Degradation of abiotically aged LDPE films containing pro-oxidant by bacterial consortium. Polym Degrad Stab 93: 1917-1922.
- Satlewal A, Soni R, Zaidi M, Shouche Y, Goel R (2008) Comparative biodegradation of HDPE and LDPE using an indigenously developed microbial consorium. J Micobiol Biotechnol 18: 477-482.
- Koutny M, Amato P, Muchova M, Ruzicka J, Delort AM (2009) Soil bacterial strains able to grow on the surface of oxidized polyethylene film containing prooxidant additives. Int Biodeterior Biodegradation 63: 354-357.
- Watanabe T, Ohtake Y, Asabe H, Murakami N, Furukawa M (2009) Biodegradability and degrading microbes of low-density Polyethylene. J Appl Polym Sci 111: 551-559.
- Chatterjee S, Roy B, Roy D, Banerjee R (2010) Enzyme-mediated biodegradation of heat treated commercial polyethylene by Staphylococcal species. Polym Degrad Stab 95: 195-200.
- Fontanella S, Bonhomme S, Koutny M, Husarova L, Brusson JM, et al. (2010) Comparison of the biodegradability of various polyethylene films containing pro-oxidant additives. Polym Degrad Stab 95: 1011-1021.
- Feng L, Wang W, Cheng J, Ren Y, Zhao G, et al. (2007) Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci U S A 104: 5602-5607.
- Van beilen JB, Li Z, Duetz WA, Smits THM, Witholt B (2003) Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci Technol 58: 427-440.
- Belhaj A, Desnoues N, Elmerich C (2002) Alkane biodegradation in Pseudomonas aeruginosa strains isolated from a polluted zone: identification of alkB and alkB-related genes. Res Microbiol 153: 339-344.
- Saadoun I, Alawawdeh M, Jaradat Z, Ababneh Q (2008) Growth of Streptomyces spp. from hydrocarbon-polluted soil on diesel and their analysis for the presence of alkane hydroxylase gene (alkB) by PCR. World J Microbiol Biotechnol 24: 2191-2198.
- Lo Piccolo L, De Pasquale C, Fodale R, Puglia AM, Quatrini P (2011) Involvement of an alkane hydroxylase of Gordonia sp. strain SoCg in degradation of solid n-alkanes. Appl Environ Microbiol 77: 1204-1213.
- Reddy MM, Deighton M, Gupta RK, Bhattacharya SN, Parthasarathy R (2009) Biodegradation of oxo-biodegradable polyethylene. J Appl Polym Sci 111: 1426-1432.
- Maier T, Förster HH, Asperger O, Hahn U (2001) Molecular Characterization of the 56-kDa CYP153 from Acinetobacter sp. EB104. Biochem Biophys Res Commun 286: 652-658.
- van Beilen JB, Funhoff EG, van Loon A, Just A, Kaysser L, et al. (2006) Cytochorome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl Environ Microbiol 72: 59-65.
- Funhoff EG, Salzmann J, Bauer U, Witholt B, van Beilen JB (2007) Hydroxylation and epoxidation reactions catalyzed by CYP153 enzymes. Enzyme Microb Technol 40: 806-812.
- Marchant R, Sharkey FH, Banat IM, Rahman TJ, Perfumo A (2006) The degradation of n-hexadecane in soil by thermophilic geobacilli. FEMS Microbiol Ecol 56: 44-54.
- Smits TH, Röthlisberger M, Witholt B, van Beilen JB (1999) Molecular screening for alkane hydroxylase genes in Gram-negative and Gram-positive strains. Environ Microbiol 1: 307-317.
- Kim MN, Park ST (2010) Degradation of poly(L-lactide) by a mesophilic bacterium. J Appl Polym Sci 117: 67-74.
- Kim MN, Yoon MG (2010) Isolation of strains degrading poly(vinyl alcohol) at high temperatures and their biodegradation ability. Polym Degrad Stab 95: 89- 93.
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680.
- Kumar S, Tamura K, Nei M (2004) MEGA 3: Integrated software for molecular evolutionary genetic analysis and sequence alignment. Brief Bioinform 5: 150- 163.
- Lee SH, Kim MN (2010) Isolation of bacteria degrading poly (butylene succinate-co-butylene adipate) and their lip A gene. Int Biodeterior Biodegradation 64: 184-190.
- Chaffin DO, Rubens CE (1998) Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219: 91-99.
- Determination of the ultimate aerobic biodegradability and disintegration of plastic materials under controlled composting conditions Part 1: Analysis of evolved carbon dioxide by titration method, Seoul, South Korea.
- Kwon SW, Kim JS, Park IC, Yoon SH, Park DH, et al. (2003) Pseudomonas koreensis sp. nov., Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov., novel species from farm soils in Korea. Int J Syst Evol Microbiol 53: 21-27.
- Poirot O, O’Toole E, Notredame C (2003) Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res 31: 3503-3506.
- Korea Coast Guard (2008) KCG2008 White Paper. Korea Coast Guard, Korean.
- Chaerun SK, Tazaki K, Asada R, Kogure K (2004) Bioremediation of coastal areas 5 years after the Nakhodka oil spill in the Sea of Japan: isolation and characterization of hydrocarbon-degrading bacteria. Environ Int 30: 911-922.
- Nowak B, Pajak J, Drozd-Bratkowicz M, Rymarz G (2011) Microorganisms participating in the biodegradation of modified polyethylene films in different soils under laboratory conditions Int Biodeterior Biodegradation 65: 757-767.
- Teeraphatpornchai T, Nakajima-Kambe T, Shigeno-Akutsu Y, Nakayama M, Nomura N, et al. (2003) Isolation and characterization of a bacterium that degrades various polyester-based biodegradable plastics. Biotechnol Lett 25: 23-28.
- Koutny M, Lemaire J, Delort AM (2006) Biodegradation of polyethylene films with prooxidant additives. Chemosphere 64: 1243-1252.
- Kohno T, Sugimoto Y, Sei K, Mori K (2002) Design of PCR Primers and gene probes for general detection of alkane-degrading bacteria. Microbes Environ 17: 114-121.
- Kloos K, Munch JC, Schloter M (2006) A new method for the detection of alkane-monooxygenase homologous genes (alkB) in soils based on PCR-hybridization. J Microbiol Methods 66: 486-496.
- Heiss-Blanquet S, Benoit Y, Maréchaux C, Monot F (2005) Assessing the role of alkane hydroxylase genotypes in environmental samples by competitive PCR. J Appl Microbiol 99: 1392-1403.
- Lo Piccolo L, De Pasquale C, Fodale R, Puglia AM, Quatrini P (2011) Involvement of an alkane hydroxylase system of Gordonia sp. Strain SoCg in degradation of solid n-alkanes. Appl Environ Microbiol 77: 1204-1213.
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 28071
- [From(publication date):
April-2012 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 21377
- PDF downloads : 6694