 |
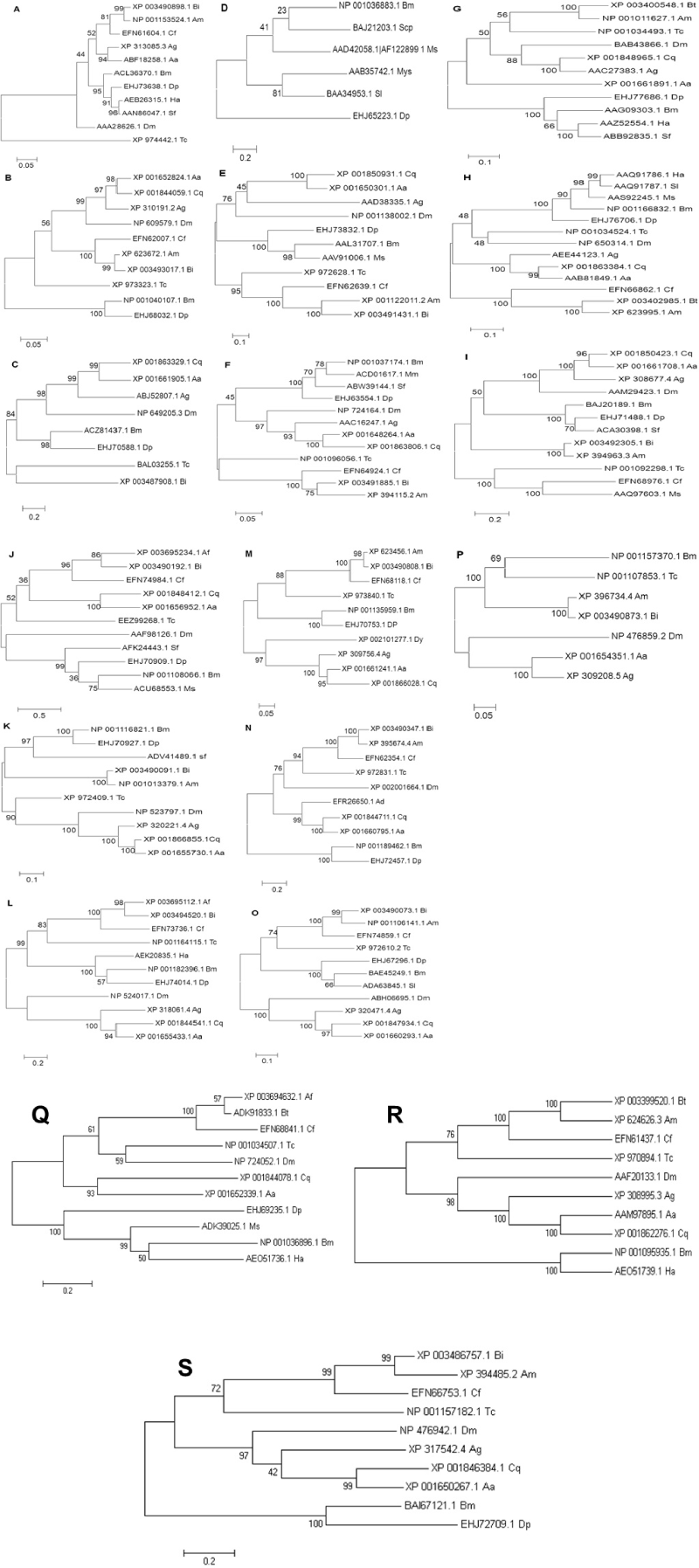
| Figure 2A-2S: Phylogram realised after performing multiple sequence alignment by CLUSTAL W using total amino acid sequence of host-response proteins that are activated in the final instar larva of B. mori and its homologues from representatives of different insect orders: The proteins analysed were (A) – Hsp70, (B) – Chaperonin subunit 4 Delta, (C) – SERPIN – 4, (D) – paralytic peptide (PP), (E) – prophenol oxidase activating enzyme (PPAE), (F) – DOPA decarboxylase (DDC), (G) – prophenol oxidase 2 (PPO2), (H) – Chitinase, (I) – ⊆-N-Acetylglucosaminidase, (J) – Spatzle, (K) – BmToll (18 wheeler), (L) – Nedd2 like Caspase, (M) – Autophagy 5- like (Atg5), (N) – Apoptosis - inducing factor (AIF), (O) – Trehalase, (P) – Notch, (Q) Dorsal, (R) – Relish and (S) – Cactus. Details of accession numbers of proteins, name of the insects and similarity exhibited are given in the Supplementary Table 2. Numbers in the nodes represent bootstrap values computed from 1000 replications. |