Research Article Open Access
Antibiotic Susceptibility and Heavy Metal Tolerance Pattern of Serratia Marcescens Isolated From Soil and Water
| Natasha Nageswaran1, P.W. Ramteke2, O.P. Verma*3 and Avantika Pande3 | |
| 1Department of Microbiology and Fermentation Technology, Jacob School of Biotechnology & Bioengineering, SHIATS Allahabad-211007, Uttar Pradesh, India | |
| 2Department of Biological Sciences, Jacob School of Biotechnology & Bioengineering, SHIATS Allahabad-211007, Uttar Pradesh, India | |
| 3Department of Molecular & Cellular Engineering, Jacob School of Biotechnology & Bioengineering, SHIATS Allahabad-211007, Uttar Pradesh, India | |
| Corresponding Author : | O.P. Verma Department of Molecular & Cellular Engineering Jacob School of Biotechnology & Bioengineering SHIATS Allahabad-211007 Uttar Pradesh, India E-mail: vermaop.aaidu@gmail.com |
| Received May 03, 2012; Accepted June 20, 2012; Published June 22, 2012 | |
| Citation: Nageswaran N, Ramteke PW, Verma OP, Pandey A (2012) Antibiotic Susceptibility and Heavy Metal Tolerance Pattern of Serratia marcescens Isolated From Soil and Water. J Bioremed Biodeg 3:158. doi: 10.4172/2155-6199.1000158 | |
| Copyright: © 2012 Nageswaran N, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
The antibiotic and metal tolerance patterns of Serratia marcescens strains isolated from soil and water around the Sangam region of Allahabad were obtained. Using the standard minimum inhibitory concentration (MIC) for each antibiotic respectively, the Kirby-Bauer disc-diffusion method was used to obtain antibiotic resistance patterns of the Serratia strains and the MIC of the metals - chromium, cadmium, cobalt, copper, lead and nickel for each of the strains were also obtained. Plasmid curing was carried out for specific antibiotic and metal resistances to ascertain plasmid-borne transfer of resistance genes. Results obtained showed that Multi-drug resistant (MDR) strains of Serratia were resistant to certain metals as well suggesting specific metal-antibiotic resistant gene patterns in the different strains.
| Keywords |
| Disc-diffusion; Minimum inhibitory concentration; Multi-drug resistance; Plasmid curing |
| Introduction |
| Microorganisms are ubiquitous in nature and are involved in almost all biological processes of life. With rapid urbanization and natural processes, heavy metals have been found in increasing proportions in microbial habitats. Metals have been known to play a major role either directly or indirectly in almost all metabolic processes, growth and development of microorganisms [1,2]. However, increasing concentrations of metals beyond tolerance levels have forced these organisms to adapt to various biological mechanisms to cope with this condition. Hence, microbes have developed mechanisms like metal efflux systems, complexation, reduction of metal ions or utilization of the metal as a terminal electron acceptor in anaerobic respiration to tolerate heavy metal accumulation [3]. Bacteria that are resistant to such heavy metals and have the ability to grow in high concentrations of these metals play an important role in their biological cycling which has great potential in bioremediation of poorly cultivable soil high in heavy metal content. Heavy metal tolerance has been observed in the Enterobacteriaceae member, Serratia marcescens and has been thought to be attributed to plasmid-borne resistant genes. 8 isolated strains of this microorganism have been used for heavy metal tolerance testing against various metal salts in order to identify specific strains that can be used for removal of particular metals from environments such as soil and water where they are present as pollutants. |
| Antibiotic susceptibility has also emerged as an ever increasing health hazard due to the indiscriminate use of antibiotics. This has led to severe complications in patients especially with gram negative bacterial infections as the number of drugs to combat this category of infections are limited. Multidrug resistance (MDR) can also be caused by another mechanism of accumulation of multiple antibiotic resistance genes each coding for a single antibiotic occurring on resistance (R) plasmids [4]. Multi Drug Resistance organisms are posing to be a huge threat in treatment procedures due to the presence of plasmid borne mobile resistance genes that can readily spread through bacterial populations and efflux systems to counter third and even fourth generation cephalosporin [5]. |
| Serratia marcescens, formerly known as Chromobacterium prodigiosin [6] is a gram-negative facultative anaerobe [7] that has emerged in recent times as a nosocomial pathogen. It functions as an opportunistic organism in immunocompromised patients because of its invasive ability to adhere to hospital instrumentation such as catheters, endoscopes and intravenous tubing [8]. Serratia marcescens is one of the major nosocomial pathogen found associated with urinary and respiratory tract infections, endocarditis, osteomyelitis, septicemia, wound infections, eye infections (conjunctivitis) and meningitis [9]. Serratia marcescens is the only pathogenic species in its genus. The organism inhabits a wide variety of ecological niches and causes diseases in plant, vertebrate as well as invertebrate hosts [10]. |
| Various metals such as cadmium, copper, chromium, cobalt, lead and nickel were used for metal tolerance tests against 8 strains of Serratia marcescens as well as antibiotic susceptibility test was conducted for the above mentioned strains using 14 different antibiotics. All the 8 strains were isolated from soil and water near the Sangam region (confluence of the Ganga and Yamuna rivers) of Allahabad. |
| All metals and antibiotics towards which the strains showed resistance (for metals at a particular concentration) were subjected to plasmid curing experiments to determine the likelihood of plasmidborne resistance pattern and relationship between heavy metal and antibiotic resistance genes. |
| Materials and Methods |
| Preliminary tests for Identification |
| All 8 strains of Serratia marcescens were identified as Gram negative bacilli by the Gram Staining technique and produced the characteristic red pigment, Prodigiosin. |
| Biochemical tests were conducted for all the above according to Bergey’s Manual of Determinative Bacteriology and all strains were confirmed as Serratia marcescens. |
| Antibiotic susceptibility test |
| The isolated strains were checked for antibiotic resistance with Ceftriaxone, Ciprofloxacin, Cloxacillin, Gentamicin, Amikacin, Cefuroxime, Netilmicin, Erythromycin, Ceftazidime, Chloramphenicol, Vancomycin, Ampicillin, Cefepime, and Imipenem [11]. |
| The Kirby-Bauer disk diffusion method [12] was employed for antibiotic susceptibility of the 8 strains of Serratia marcescens [13]. The strains of Serratia were inoculated into 5 ml of Nutrient broth and incubated overnight at 37°C. These pure broth cultures were then swabbed on 20 ml of Mueller Hinton pre-made agar plates each. All 14 antibiotic disks were placed on each of the swabbed plates at appropriate distances from one another and the plates were then incubated at 37°C for another 24 hrs. Zones of inhibition were obtained by measuring the diameter across the centre of each zone in millimeters. |
| In addition to this, powdered form of other antibiotics were also used for antibiotic sensitivity tests such as Ampicillin, Ceftazidime, Cefepime, Cephalexin, Nalidixic acid, Neomycin Sulphate, Polymyxin, Tetracycline, Cephataxime, Streptomycin and Trimethoprim. |
| Bacterial isolation to obtain pure culture |
| Nutrient agar medium was prepared by adding sodium chloride (5.0 g), peptone (5.0 g), beef extract (3.0 g) and agar (15.0 g) to 1000 ml of distilled water and at pH 7.0 and autoclaved at 15 psi. |
| Isolates of Serratia marcescens were plated on nutrient agar and incubated at 37°C for 24 hours. The pure cultures obtained were inoculated into tubes containing 5 ml of sterile nutrient broth each and incubated overnight at 37°C. |
| Preparation of metal stock solutions |
| 1. Metal salts were used to prepare stock solutions of 100 μg/ml and 1000 μg/ml (for further tests) |
| 2. Weight of the salt required to make stock solutions is calculated [14] in the following table 1. |
| Minimum inhibitory concentration (MIC) of heavy metals |
| The isolated bacterial cultures were checked for their respective MICs towards heavy metals ions such as chromium (Cr), cadmium (Cd), Lead (Pb), copper (Cu), cobalt (Co) and nickel (Ni). The isolated pure cultures were plated on NA medium containing 3 different concentrations of each of the metals- 25 μg/ml, 50 μg/ml and 100 μg/ ml respectively. NA medium was prepared by the above mentioned method. 20 ml of NA was poured into each Petri plate and appropriate volume of metal stock solutions were pipetted into the plate. The molten NA was mixed with the metal solution by swirling the plate and was allowed to solidify. |
| The isolated cultures were streaked onto the NA medium containing metal salts using sterile loops and then incubated at 37°C for 24-48 hrs. The plates were checked for bacterial growth. The concentration of metal where there was no growth is observed as the MIC for that salt for that strain [14]. The isolates, for which growth was visible even at 100 μg/ml for a particular metal, were further grown at higher metal concentration ranges of 200 μg/ml, 400 μg/ml and 800 μg/ml respectively from a 1000 μg/ml metal stock solution. These were then incubated at 37°C for 24-48 hrs and checked for visible growth. |
| Plasmid curing |
| Each strain was checked for their resistance towards a particular antibiotic and metal. For metals, the highest concentration which showed growth of a particular strain was taken for plasmid curing. The antibiotics and metals that were used for curing were selected after MIC of metals and antibiotic resistance patterns were obtained as shown in the following Table 2. For Plasmid Curing, the protocol followed was in accordance to [15]. 200 μg/ml of acridine orange solution (mutagenic agent) was prepared in distilled water. 9 ml of peptone water and 10 ml of acridine orange solution were autoclaved separately. 9 ml of peptone water was mixed with 1 ml acridine orange solution. In another tube, 5 ml of peptone water was inoculated with Serratia marcescens culture at 37°C in the morning. After about 7-8 hrs, 0.1 ml of this broth was transferred to a fresh 10 ml solution of acridine orange and incubated at 37°C for 24 hrs under shaking conditions. Appropriate dilutions were plated on LB agar to obtain about 120 colonies. These colonies were picked aseptically and transferred on Antibiotic and metal infused plates of LB agar separately towards which the organism showed resistance earlier. The colonies which grew on the plate were those which still showed resistance and those which were cured, failed to grow on the plate since they were plasmid borne genes and were sensitive to the particular metal or antibiotic. |
| Results and Discussion |
| All the eight isolates of Serratia marcescens that were obtained from the soil and river water samples of the Sangam region (confluence of the Ganga and Yamuna rivers) at Allahabad produced the characteristic red pigment, prodigiosin. Their antibiotic susceptibility and tolerance to heavy metals are shown in Table 3 & 4 respectively. All strains were sensitive towards Ceftriaxone, Chloramphenicol, Vancomycin, Netilmicin, Erythromycin, Ciprofloxacin, Cefuroxime, Amikacin, Gentamicin, Cloxacillin, Neomycin sulphate, Polymyxin, Tetracycline, and Cephataxime. All strains were sensitive towards Imipenem, a β-lactam Carbapenem [16] except for Strain 5. Imipenem is a high end broad spectrum drug and is used to combat very severe gram positive and gram negative infections especially nosocomial infections. |
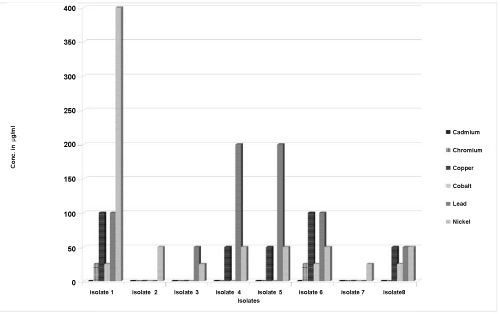
| The isolates were further tested for heavy metal tolerance against six heavy metal compounds in order to study their ability to grow in the presence of increasing concentrations of respective heavy metals as shown in Figure 1. Cadmium proved to be toxic to all the isolates of Serratia even at low concentrations of 25 μg/ml. Isolate one grew in Chromium and Cobalt at 25 μg/ml but was inhibited at 50 μg/ ml respectively. It grew at higher concentrations of 100 μg/ml for Copper, Lead and Nickel. Isolate 2 did not grow in any of the metals except Nickel at 50 μg/ml and was inhibited at 100 μg/ml. Isolate 3 showed visible growth for Lead and Nickel at 50 μg/ml and 25 μg/ml respectively but did not grow in the other metal compounds. Isolate 4 and 5 showed identical tolerances to 50 μg/ml of Copper and Nickel and 100 μg/ml of Lead. Both did not grow on Chromium and Cobalt. Isolate 6 tolerated 100 μg/ml of Copper and Lead, 50 μg/ml of Nickel and 25 μg/ml of Chromium and Cobalt. Isolate 7 could only tolerate 25 μg/ml of Nickel. Isolate 8 was able to tolerate 50 μg/ml of Copper, Lead and Nickel and 25 μg/ml of cobalt. |
| All the isolates which grew at and below 100 μg/ml concentration of a particular metal were further tested at higher concentrations of 200 μg/ml, 400 μg/ml and 800 μg/ml of the respective metals in order to establish the concentration at which the concerned isolate was inhibited for that particular metal. Only Isolate 1 grew at 400 μg/ml concentration of Nickel and Isolates 4 and 5 were able to tolerate up to 200 μg/ml concentration of Lead. Plasmid curing studies showed variable results for all the strains of Serratia marcescens revealing that they behaved differently in their plasmid borne resistant gene patterns. |
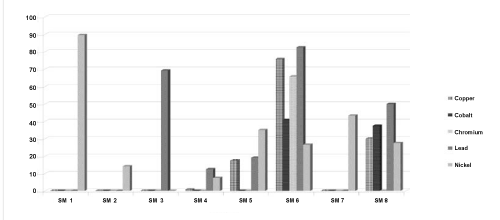
| Isolate 1 showed about 90% curing for Nickel alone. Isolate 3 showed about 69% curing for Lead. Isolate 6 revealed 76% curing for Copper, 66% curing for Chromium and 83% curing for Lead. The rest of the isolates did not give significant curing results. The above results have been graphically represented in Figure 2. |
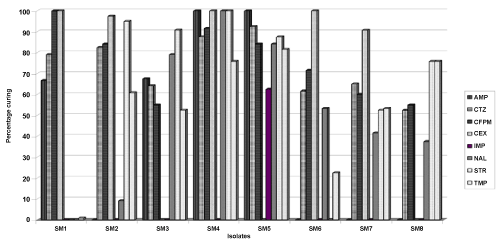
| Isolate 1 showed moderate levels of curing for ampicillin and ceftazidime and absolute curing for cefepime and cephalexin. Isolate 2 showed significantly high levels of curing for ceftazidime, cefepime, cephalexin and streptomycin and 60% curing for trimethoprim. Isolate 3 showed moderate levels of curing for ampicillin, ceftazidime, and nalidixic acid and 90% for streptomycin. Isolate 4 showed very high levels of curing for nearly all antibiotics. Results obtained showed 100% curing for ampicillin, cephalexin, nalidixic acid and streptomycin. It also showed 87% curing for ceftazidime, 91% for cefepime and 76% for trimethoprim. Isolate 5 showed very high levels of curing for ampicillin, ceftazidime, cefepime, nalidixic acid, streptomycin and trimethoprim and moderate levels of curing for imipenem. Isolate 6 showed reasonable levels of curing for ceftazidime and cefepime and 100% curing for cephalexin. Isolate 7 moderate levels of curing for ceftazidime and cefepime and high levels of curing for cephalexin. Isolate 8 showed reasonable levels of curing for only streptomycin and trimethoprim (Figure 3). |
| For each strain, percentage curing was carried out numerically towards the different antibiotics and metals, and those which showed curing beyond 50% were said to have been significantly cured as seen in Table 4. |
| Environments containing elevated concentrations of heavy metals potentially give rise to heavy metal tolerant organisms [17]. The area around the Sangam region of Allahabad, Uttar Pradesh, India has shown a large variety of microorganisms out of which Serratia marcescens has shown significant antibiotic and heavy metal resistant patterns. In order to survive these metal stressed conditions, microorganisms have evolved to tolerate the uptake of heavy metal ions [18]. Different strains of Serratia have shown varied resistant patterns suggestive of plasmidborne transfer of resistant genes or pump effluxing systems. However, it has been suggested that increasing antibiotic resistance of Gramnegative bacteria is mainly due to mobile genes on plasmids that can readily travel through bacterial populations [5]. The studies conducted for Serratia show evidence of resistant genes in accordance to [4]. All Serratia strains that were tolerant to metals were also tolerant towards several antibiotics also seen by [19] who studied bacteria isolated from drinking water that contained organisms that were metal as well as antibiotic resistant. This has significance suggesting that the metal and antibiotic resistances are a function. |
| Conclusion |
| The above studies have shown the dual properties of the bacterial genome in rendering the metal tolerant organisms antibiotic resistant as well. Further research in closely examining this interesting nature of the Serratia genome will enable us to find out the exact mechanism of these two shared properties and their close association with one another. The metal tolerant nature of Serratia marcescens has tremendous potential in the bioremediation of heavy metal accumulation in soil and water and also in the treatment of sewage and toxic wastes. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 16514
- [From(publication date):
July-2012 - Nov 03, 2025] - Breakdown by view type
- HTML page views : 11500
- PDF downloads : 5014
