Research Article Open Access
Analyzing Interaction of μ-, δ- and κ-opioid Receptor Gene Variants on Alcohol or Drug Dependence Using a Pattern Discovery-based Method
Zhong Li1 and Huiping Zhang2,3*1Department of Computational Genetics, High Throughput Biology Inc., Summit, NJ, USA
2Department of Psychiatry, Yale University School of Medicine, New Haven, CT, USA
3VA Connecticut Healthcare System, West Haven Campus, CT, USA
- *Corresponding Author:
- Huiping Zhang
Department of Psychiatry
Yale University School of Medicine
VA Medical Center/116A2, 950 Campbell Avenue
West Haven, CT 06516, USA
Tel: (203)932-5711 ext. 5245
Fax: (203)937-4741
E-mail: huiping.zhang@yale.edu
Received April 08, 2013; Accepted May 03, 2013; Published May 14, 2013
Citation: Li Z, Zhang H (2013) Analyzing Interaction of µ-, δ- and κ-opioid Receptor Gene Variants on Alcohol or Drug Dependence Using a Pattern Discovery-based Method. J Addict Res Ther S7:007. doi:10.4172/2155-6105.S7-007
Copyright: © 2013 Li Z, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Addiction Research & Therapy
Abstract
Background: Polymorphisms in the μ-, δ- and κ-opioid receptor genes (OPRM1, OPRD1 and OPRK1) have been reported to be associated with substance (alcohol or drug) dependence. The influence of an individual gene on a disease trait should be more evident when analyzed in the context of gene-gene interactions. Thus, we assessed the joint effect of variants in these three opioid receptor genes on alcohol, cocaine, or opioid dependence.
Methods: Genotype data for 13 OPRM1 Single Nucleotide Polymorphisms (SNPs), 11 OPRD1 SNPs and seven OPRK1 SNPs were obtained from 382 European Americans (EAs) affected with substance dependence [among them, 318 with Alcohol Dependence (AD), 171 with Cocaine Dependence (CD), and 91 with Opioid Dependence (OD)] and 338 EA control subjects. We assessed the joint effect of OPRM1, OPRD1 and OPRK1 variants on AD, CD, or OD using a pattern discovery-based association test. Specific marker patterns (consisting of alleles of OPRM1, OPRD1 and OPRK1) that were significantly more frequent in AD, CD, or OD cases than in controls were identified.
Results: 12 significant patterns in the AD dataset, four significant patterns in the CD dataset, and 18 significant patterns in the OD dataset were identified. Moreover, the significance of most marker patterns was due primarily to OPRM1 variants and, to a lesser degree, OPRD1 variants.
Conclusion: Our findings suggest that variation in the above three opioid receptor genes can jointly influence the vulnerability of individuals to alcohol or drug dependence. Evidence provided by this study also supports previous biological findings that the interaction of the three opioid receptors can modulate the action of opioid and non-opioid drugs and alcohol.
Keywords
Opioid receptor genes; Case-control genetic association study; Gene-gene interaction; Pattern discovery-based association test
Introduction
Substance dependence, such as alcohol, cocaine, or opioid dependence, is a set of genetically complex disorders due to the effect of a number of different individual disease genes (heterogeneity) or a combination of different disease genes (polygeneity). In addition, environmental factors also have a strong influence on the development of substance dependence. Given the high rate of co-morbidity of alcohol, cocaine, and opioid dependence, and consistent with studies in genetic epidemiology, it is likely that, besides specific genetic factors that are responsible for each of the substances abused, common genetic factors may be involved in these disorders as well [1,2]. There is evidence that the three opioid receptor genes (OPRM1, at 6q24-q25, which encodes the μ-opioid receptor; OPRD1, at 1p36.1-p34.3, which encodes the δ-opioid receptor; and OPRK1, at 8q11.2, which encodes the κ-opioid receptor) could be such common genetic factors [3-6].
The above three opioid receptors are the molecular targets for endogenous opioid peptides, opioid analgesic agents, and commonly abused opioid drugs like heroin. There is mounting evidence that the three receptors directly mediate reward, tolerance, and dependence associated with opioids [7,8]. They are also indirectly involved in the reinforcing properties of non-opioid drugs (such as cocaine and alcohol) due to the intimate relationship between the opioid system and the mesolimbic dopamine system. Dopamine is known to be a key neurotransmitter interacting with the brain reward center [9,10]. Cocaine binds to the dopamine transporter and inhibits dopamine re-uptake in the Nucleus Accumbens (NAc), thus increasing synaptic dopamine levels and stimulating dopaminergic transmission [11,12], whereas ethanol directly stimulates dopaminergic neurons in the Ventral Tegmental Area (VTA), leading to increased release of dopamine in the NAc [13]. The basal dopamine level in the dopamine system is under the tonic control of two opposing opioid systems: activation of the μ-receptor (and possibly the δ-receptor) in the VTA increases extracellular dopamine levels in the NAc; activation of the κ-receptor in the VTA decreases extracellular dopamine levels in the NAc [14,15].
Additionally, interaction of the three opioid receptors can modulate the action of opioid and non-opioid drugs and alcohol. There is evidence of physical and functional interactions between μ- and δ-opioid receptors. Extensive co-localization of μ- and δ-receptors has been observed in brain reward regions [16-18]. The apposition of μ- and δ-receptors suggests that these two receptors are functionally interrelated. Several studies have demonstrated modulatory interactions between μ- and -receptors. For example, δ-agonists can enhance the analgesic potency and efficacy of μ agonists (e.g., morphine), and δ-antagonists can prevent or diminish the development of tolerance and physical dependence by μ agonists [19,20]. Of interest, μ-δ heterodimers, which exhibit ligand binding and signalling characteristics distinct from those of μ- and δ-receptors, have been isolated from cells co-expressing these two receptors [21]. In contrast to the interaction between μ- and δ-receptors, opposing interactions have been observed between μ- and κ-receptors. Activation of the κ-receptor by κ-receptor agonists opposed a variety of μ-receptor mediated actions in the brain, including analgesia, tolerance, reward, and memory processes [22]. However, the inhibitory effect of κ-receptor agonists on the function of the μ-receptor can be completely reversed by the κ-receptor antagonist nor-BNI [23]. Similarly, opposing interactions have been observed between δ- and κ-opioid receptors. In addition, δ- and κ-receptors can form heterodimers that exhibit ligand binding and functional properties that are different from those of either receptor. The δ-κ heterodimer can bind highly selective agonists and potentiate signal transduction [24].
Considering the close biological interaction of the three receptors, we hypothesized that variation in their genes (OPRM1, OPRD1, and OPRK1) might have joint effects on risk for alcohol or drug dependence. We sought to test this hypothesis via a powerful multi-locus analysis method based on an efficient pattern discovery algorithm.
Materials and Methods
Sample and genotype data
As described in our two previous studies [4,6], genotype data of 13 OPRM1 SNPs, 11 OPRD1 SNPs, and seven OPRK1 SNPs were obtained from 376 European American cases (280 males and 96 females), who met lifetime DSM-III-R (American Psychiatric Association, 1987) or DSM-IV (American Psychiatric Association, 1994) criteria for the diagnosis of alcohol, cocaine, or opioid dependence (AD, CD, or OD), and 384 European American healthy controls (143 males and 241 females). Among 376 cases, 318 were affected with AD, 166 were affected with CD, and 91 were affected with OD, respectively. The study subjects were recruited at the University of Connecticut Health Center or the VA Connecticut Healthcare System-West Haven Campus. The study protocol was approved by the Institutional Review Board (IRB) at each clinical site. Informed consent was obtained from participants before they entered the study, and they were paid for their participation.
Multi-locus interaction and disease association analyses
The Pattern Examiner program [25,26], which is a pattern discovery-based association analysis approach, was applied in analyzing the interactive effect of variation in three opioid receptor genes (OPRM1, OPRD1, and OPRK1) on alcohol or drug dependence. Pattern Examiner is a non-parametric data mining method for detection of multi-locus gene-gene or gene-environment interactions in population-based case-control studies. This method has two major steps: (1) pattern discovery, and (2) significance evaluation. Briefly, data are organized in a two-dimensional matrix with markers as columns, individuals as rows, and individuals’ alleles or genotypes as cell values. Each marker is represented by five columns: two for each of the two alleles and three for each of the three possible genotypes. A pattern is defined as a maximal sub-matrix of the data matrix in which the value of each marker across all individuals in the sub-matrix satisfies a predefined equivalence criterion such as the same genotype value. A sub-matrix is maximal if (1) no more rows can be added while keeping the columns fixed, and (2) no more columns can be added while keeping the rows fixed. Under this formulation, patterns can be used to model both multi-locus allelic and multi-locus genotypic contributions to a disease state. In the pattern discovery step, patterns are identified using input data from the case population alone to uncover elevated risk factors enriched in the case population (patterns from the control population alone can also be examined to uncover protective factors). The extensiveness and execution time of the pattern discovery step are controlled by two parameters: the support threshold, which specifies the minimum number of rows a pattern must have; and the locus threshold, which specifies the extent of locus interaction. For example, with the support and locus thresholds set to 20 and 2, respectively, all reported patterns will have 20 or more case supports and mostly one or two markers. In the significance evaluation step, a 2×2 contingency table is constructed for each pattern to tally its support in the case and control populations (“case support” and “control support,” respectively). The two categorical variables tabulated are Population Type (“cases” vs. “controls”) and Pattern Match Status (“matches” vs. “does not match”). Partially missing data are excluded. p values are obtained from a ᵠ2 test of independence and then adjusted for multiple testing.
To examine the interaction of OPRM1, OPRD1, and OPRK1, we performed a two-locus marker-based gene-gene interaction analysis using the above Pattern Examiner algorithm. The support threshold and the locus threshold parameters were set to 1 and 2, respectively, meaning that all reported patterns had at least one case support and no more than two loci were included in the analysis. The support threshold of 1 was chosen so that all possible patterns were identified and evaluated. A modified Bonferroni correction for multiple testing was applied to generate the adjusted p values for the identified patterns. The correction factor for the modified Bonferroni correction was the total number of patterns that contained equal or greater case support than the target pattern rather than the total number of patterns identified. As a result, the modified Bonferroni correction was less stringent than the direct Bonferroni correction (in which the correction factor was the total number of patterns identified) and thus yielded fewer false negatives. In both multiple testing correction schemes, the adjusted significance was robust against the arbitrary selection of values for the support threshold parameter in the pattern discovery step. The odds ratio with its confidence interval was also calculated for each pattern.
| SNP ID (this study) | SNP ID (NCBI rs#) | Gene | Alleles | MAFa | Location | Amino Acid Change | Chromosome Position (bp) | Genotyping Method |
|---|---|---|---|---|---|---|---|---|
| M1 | rs1799971 | OPRM1 | A118G | 0.13 (G) | Exon 1 | Asn40Asp | Chr06: 154402490 | PCR-RFLP |
| M2 | rs511435 | OPRM1 | A/G | 0.21 (A) | Intron 1 | - | Chr06: 154410240 | TaqMan |
| M3 | rs524731 | OPRM1 | A/C | 0.20 (A) | Intron 1 | - | Chr06: 154416785 | TaqMan |
| M4 | rs3823010 | OPRM1 | A/G | 0.15 (A) | Intron 1 | - | Chr06: 154420845 | TaqMan |
| M5 | rs495491 | OPRM1 | C/T | 0.24 (C) | Intron 1 | - | Chr06: 154424235 | TaqMan |
| M6 | rs1381376 | OPRM1 | A/G | 0.15 (A) | Intron 1 | - | Chr06: 154434951 | TaqMan |
| M7 | rs3778156 | OPRM1 | A/G | 0.15 (G) | Intron 1 | - | Chr06: 154446006 | TaqMan |
| M8 | rs2075572 | OPRM1 | C/G | 0.43 (G) | Intron 2 | - | Chr06: 154453697 | TaqMan |
| M9 | rs548646 | OPRM1 | C/T | 0.34 (T) | Intron 3 | - | Chr06: 154459840 | TaqMan |
| M10 | rs9322447 | OPRM1 | A/G | 0.48 (A) | Intron 3 | - | Chr06: 154466013 | TaqMan |
| M11 | rs609148 | OPRM1 | C/T | 0.25 (T) | Intron 3 | - | Chr06: 154472707 | TaqMan |
| M12 | rs648893 | OPRM1 | C/T | 0.25 (C) | Intron 3 | - | Chr06: 154480321 | TaqMan |
| M13 | rs671531 | OPRM1 | A/G | 0.35 (A) | downstream | Chr06: 154482434 | TaqMan | |
| D1 | rs569356 | OPRD1 | C/T | 0.12 (C) | upstream | Chr01: 29009273 | PCR-RFLP | |
| D2 | rs1042114 | OPRD1 | G80T | 0.13 (G) | exon 1 | Cys27Phe | Chr01: 29011562 | PCR-RFLP |
| D3 | rs678849 | OPRD1 | C/T | 0.47 (C) | intron 1 | - | Chr01: 29017775 | PCR-RFLP |
| D4 | rs2236857 | OPRD1 | A/G | 0.30 (G) | intron 1 | - | Chr01: 29034196 | TaqMan |
| D5 | rs2236855 | OPRD1 | G/T | 0.29 (T) | intron 1 | - | Chr01: 29034586 | TaqMan |
| D6 | rs2298896 | OPRD1 | A/C | 0.37 (C) | intron 1 | - | Chr01: 29038725 | TaqMan |
| D7 | rs421300 | OPRD1 | C/T | 0.38 (C) | intron 1 | - | Chr01: 29042180 | TaqMan |
| D8 | rs529520 | OPRD1 | G/T | 0.50 (T) | intron 1 | - | Chr01: 29047533 | TaqMan |
| D9 | rs12749204 | OPRD1 | A/G | 0.21 (G) | intron 1 | - | Chr01: 29048800 | TaqMan |
| D10 | rs2234918 | OPRD1 | C921T | 0.44 (C) | exon 3 | Gly307Gly | Chr01: 29062184 | PCR-RFLP |
| D11 | rs204076 | OPRD1 | A/T | 0.34 (A) | downstream | Chr01: 29062977 | TaqMan | |
| K1 | rs12675595 | OPRK1 | A/G | 0.09 (A) | upstream | Chr08: 54330478 | TaqMan | |
| K2 | rs1051660 | OPRK1 | G36T | 0.10 (T) | exon 1 | Pro12Pro | Chr08: 54326115 | TaqMan |
| K3 | rs6985606 | OPRK1 | C/T | 0.46 (T) | intron 1 | - | Chr08: 54323669 | TaqMan |
| K4 | rs997917 | OPRK1 | C/T | 0.34 (C) | intron 1 | - | Chr08: 54314931 | TaqMan |
| K5 | rs702764 | OPRK1 | C843T | 0.12 (C) | exon 3 | Ala281Ala | Chr08: 54304710 | TaqMan |
| K6 | rs963549 | OPRK1 | C/T | 0.14 (T) | exon 3 (UTR) | Chr08: 54304377 | PCR-RFLP | |
| K7 | rs7820807 | OPRK1 | C/T | 0.12 (C) | downstream | Chr08: 54301414 | TaqMan |
aMarker minor allele frequency (MAF) in European American (EA) healthy control subjects.
Table 1: Information and genotyping methods for OPRM1, OPRD1and OPRK1 Single Nucleotide Polymorphisms (SNPs).
Results
In our previous studies, we examined the association of 13 OPRM1 SNPs, 11 OPRD1, and seven OPRK1 SNPs (marker information is given in Table 1) with alcohol or drug dependence. Single marker and haplotype analyses have shown a positive association of variation in the three opioid receptor genes and alcohol or drug dependence [4,6]. In the present study, we further analyzed the interactive effect of OPRM1, OPRD1, and OPRK1 variants on alcohol or drug dependence using a pattern discovery-based method.
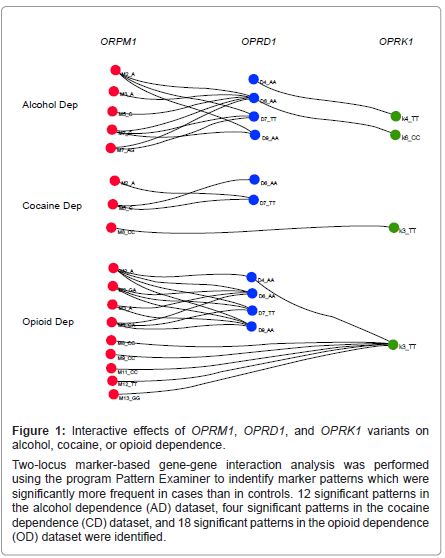
Two-locus marker-based gene-gene interaction analysis results are presented in Table 2. A small proportion of marker patterns [1.06% (12 of 1,134), 0.35% (4 of 1,147), and 1.86% (18 of 965) for alcohol, cocaine and opioid datasets, respectively] were found significantly more frequent in cases than in controls (p<0.05, after the adjusted Bonferroni correction). Figure 1 illustrates significant interactions among markers of OPRM1, OPRD1, and OPRK1. Each node in the graph represented a marker with a particular allele or genotype that was found in significant patterns. Each edge represented a significant pattern (p values were labelled on edges). The majority of the significant patterns were comprised of marker alleles of OPRM1 and OPRD1, suggesting a greater impact of these two genes on alcohol, cocaine, or opioid dependence in comparison to that of OPRK1. Remarkably, one OPRM1 SNP (M2) and two OPRD1 SNPs (D6 and D7) were consistently present in significant patterns for all three substance dependence datasets, suggesting the existence of common disease variants or a combination of common disease variants for alcohol, cocaine, and opioid dependence. Moreover, several significant marker patterns appeared in two or all three datasets. The interaction of M2_A and D7_TT was noticed in all three datasets. Additionally, patterns M2_A~D6_AA, M2_A~D9_AA, and M3_ A~D6_AA were shown in both alcohol and opioid dependence datasets, pattern M5_C~D6_AA was observed in both alcohol and cocaine dependence datasets, and pattern M8_CC~K3_TT was found in both cocaine and opioid dependence datasets. Interestingly, all markers of OPRD1 and OPRK1 in significant patterns contained homozygous genotypes, suggesting a recessive effect by OPRD1 or OPRK1 towards the disease etiology of alcohol, cocaine, or opioid dependence. A lesser consistent effect was observed for markers of OPRM1.
Discussion
To our knowledge, this is the first study to look at the joint effect of the three opioid receptor genes on three complex substance dependence traits (alcohol, cocaine, and opioid dependence) that cooccur frequently. The gene-gene interaction results support the findings in our previous single gene studies [4,6]. The significance of most marker patterns (as presented in Table 2 and Figure 1) was due primarily to OPRM1 markers and, to a lesser degree, OPRD1 markers. A plausible explanation for this finding is that, among the three receptor genes, OPRM1 variants produce the strongest effect on substance dependence; OPRD1 variants combine with OPRM1 variants to generate an additive (or possibly synergistic) effect; and OPRK1 variants can modulate the effects of OPRM1 or OPRD1 variants. The weaker role of OPRK1 variants in substance dependence observed in this study agrees with the findings in previous neuropsychopharmacological studies that the κ-opioid receptor seemed to mediate psychotomimetic effects [27], which do not have a clear relation to risk of substance dependence. In contrast, the μ-receptor (coded by OPRM1), in particular, and the δ-receptor (coded by OPRD1) to a lesser degree, play a major role in opioid drug reward and addiction.
| Dataset | Num. of cases with/without a pattern | Num. of controls with/without a pattern | Unadjusted P value | Adjusted P value | Odds Ratio (Confidence Interval) | Marker patterns |
|---|---|---|---|---|---|---|
| Alcohol Dependence (12 patterns) | 71/207 | 37/268 | 3.19×10-5 | 0.007 | 2.48 (1.60-3.85) | M5_C + D6_AA |
| 89/181 | 57/247 | 1.00×10-4 | 0.013 | 2.11 (1.44-3.11) | D4_AA + K4_TT | |
| 62/215 | 31/276 | 5.13×10-5 | 0.014 | 2.57 (1.61-4.09) | M2_A + D6_AA | |
| 60/218 | 29/277 | 4.87×10-5 | 0.014 | 2.63 (1.63-4.23) | M3_A + D6_AA | |
| 109/163 | 75/215 | 3.00×10-4 | 0.024 | 1.92 (1.34-2.74) | D6_AA + K6_CC | |
| 76/200 | 45/259 | 2.00×10-4 | 0.03 | 2.19 (1.45-3.30) | M7_G + D9_AA | |
| 46/232 | 19/285 | 8.25×10-5 | 0.032 | 2.97 (1.70-5.21) | M7_AG + D6_AA | |
| 60/216 | 31/276 | 1.00×10-4 | 0.034 | 2.47 (1.55-3.95) | M2_A + D7_TT | |
| 87/188 | 57/250 | 3.00×10-4 | 0.034 | 2.03 (1.38-2.98) | M2_A + D9_AA | |
| 52/225 | 24/280 | 1.00×10-5 | 0.038 | 2.70 (1.61-4.51) | M7_G + D7_TT | |
| 52/226 | 24/280 | 1.00×10-4 | 0.04 | 2.68 (1.60-4.49) | M7_G + D6_AA | |
| 45/232 | 19/285 | 1.00×10-4 | 0.049 | 2.91 (1.66-5.11) | M7_AG + D7_TT | |
| Cocaine Dependence (4 patterns) | 39/102 | 37/268 | 5.13×10-5 | 0.001 | 2.77 (1.67-4.59) | M5_C + D6_AA |
| 37/103 | 37/268 | 2.00×10-4 | 0.037 | 2.60 (1.56-4.33) | M5_C + D7_TT | |
| 33/107 | 31/276 | 2.00×10-4 | 0.043 | 2.75 (1.60-4.71) | M2_A + D7_TT | |
| 19/120 | 11/293 | 9.67×10-5 | 0.047 | 4.22 (1.95-9.13) | M8_CC + K3_TT | |
| Opioid Dependence (18 patterns) | 33/44 | 57/249 | 7.74×10-6 | 0.0004 | 3.28 (1.92-5.60) | M2_A + D9_AA |
| 20/58 | 24/282 | 1.06×10-5 | 0.002 | 4.05 (2.10-7.82) | M2_GA + D6_AA | |
| 22/56 | 30/275 | 2.45×10-5 | 0.005 | 3.60 (1.94-6.70) | M11_CC + K3_TT | |
| 22/55 | 31/275 | 2.87×10-5 | 0.005 | 3.55 (1.88-6.46) | M12_TT + K3_TT | |
| 30/47 | 55/250 | 8.25×10-5 | 0.005 | 2.90 (1.69-4.99) | M3_A + D9_AA | |
| 13/63 | 11/292 | 1.70×10-5 | 0.006 | 5.48 (2.35-12.79) | M8_CC + K3_TT | |
| 22/56 | 31/275 | 3.74×10-5 | 0.007 | 3.49 (1.88-6.46) | M2_A + D6_AA | |
| 27/50 | 47/259 | 9.17×10-5 | 0.01 | 2.98 (1.70-5.22) | M2_GA+ D9_AA | |
| 19/59 | 24/281 | 3.94×10-5 | 0.01 | 3.77 (1.94-7.33) | M3_CA + D6_AA | |
| 18/60 | 22/284 | 4.15×10-5 | 0.012 | 3.87 (1.96-7.66) | M13_GG + K3_TT | |
| 19/59 | 25/281 | 6.33×10-5 | 0.016 | 3.62 (1.87-7.00) | M2_GA + D7_AA | |
| 20/58 | 28/278 | 8.70×10-5 | 0.02 | 3.42 (1.81-6.49) | D4_AA + K3_TT | |
| 18/60 | 23/282 | 7.83×10-5 | 0.022 | 3.68 (1.87-7.24) | M3_CA + D7_TT | |
| 26/51 | 46/259 | 2.00×10-4 | 0.023 | 2.87 (1.63-5.06) | M3_CA+ D9_AA | |
| 21/57 | 31/275 | 1.00×10-4 | 0.024 | 3.27 (1.75-6.09) | M2_A + D7_TT | |
| 18/60 | 24/282 | 1.00×10-4 | 0.034 | 3.52 (1.80-6.90) | M9_CC + K3_TT | |
| 20/58 | 29/276 | 1.00×10-4 | 0.035 | 3.28 (1.74-6.20) | M3_A + D6_AA | |
| 25/53 | 44/262 | 3.00×10-4 | 0.041 | 2.81 (1.58-4.98) | M2_A + D4_AA |
Dataset: genotype data of controls and alcohol, cocaine, or opioid dependent cases.
Table 2: Significant marker patterns identified in two-locus marker-based gene-gene interaction analyses.
In comparison to conventional single gene/marker association analysis, multi-locus association analysis may be more powerful. Single gene/marker analysis has been widely used to study many complex disorders. However, the findings are often inconsistent. The inconsistency may be due to insufficient sample size, population stratification, random variation, and confounding factors. The multifactorial nature of complex disorders may also lead to inconsistent results. The interaction of several genes involved in a disease may complicate the findings. Although synergistic effects of multiple genes can be expected to augment the phenotypic expression of a disorder, in certain circumstances, the effect of one gene may be suppressed or opposed by another gene, and as a result, the influence of one gene in a disease may be rendered undetectable. In view of this, if the information concerning the interactive effect of genes is considered, the chance to detect the risk effect of a susceptibility locus will be increased, even when the sample size is moderate [28]. One more advantage of the pattern discovery-based association test is that, since both alleles and genotypes were included in the pattern search, it had the potential to reveal the mode of inheritance at each locus. In the two-locus marker-based analysis, a dominant or recessive mode of inheritance was seen for almost all significant patterns (Figure 1). For example, several markers of OPRM1 in significant patterns showed a dominant effect as indicated by the inclusion of a single allele. On the contrary, several other markers of OPRM1 and all markers of OPRD1 and OPRK1 consistently showed a recessive effect as indicated by the inclusion of homozygous genotypes in the significant patterns.
There are two major challenges to the use of multi-locus association analysis. One challenge is the combinatorial nature of gene-gene interaction analyses. Increasing the number of loci results in exponential growth of possible multi-locus combinations, thus leading to strenuous computation. The pattern discovery approach has been proved to be an efficient way to deal with the combinatorial nature of gene-gene interaction analyses [25]. Another challenge is how to adjust the statistical significance for multiple testing. Multi-locus combinations and strong correlations among different marker combinations due to locus sharing make the use of Bonferroni correction inappropriate. Here, we employed a modified Bonferroni correction scheme, in which the raw P value for a specific pattern was adjusted by the total number of patterns that have the same or more case support than this pattern instead of using all patterns identified (many of which may have lesser case support than this pattern). The validity of this multiple testing correction method was confirmed by Li et al. [25] using a Monte Carlo process [29,30].
In summary, the present study, by using a pattern discovery-based association test approach, has demonstrated a potential interactive effect of the three opioid receptor genes on substance dependence. Our data have shown the importance of assessing joint effects of multiple related genes on the susceptibility to complex disorders such as substance dependence. The disease association patterns identified in this study may be useful for diagnosis and prediction of substance dependence. Furthermore, these findings have important pharmacogenetic implications relevant to the treatment of substance dependence, which we would argue that the joint effect of the three opioid receptor genes must be taken into consideration.
Software Information
Issues related the software “Pattern Examiner” and data analyses performed in this report using “Pattern Examiner” can be addressed to: Zhong Li, Ph.D., High Throughput Biology Inc., 55 Union Place, Suite 126, Summit, NJ 07901, Tel: (973) 992-6222, E-mail: zli@htbiology.com
Acknowledgments
This work was supported by the Pathway to Independence Award (DA022891) from the National Institute on Drug Abuse (HZ). It was also partially supported by the Small Business Innovative Research (SBIR) phase II grant (R44 CA101432) awarded by the National Cancer Institute (Z L). In addition, the authors acknowledge support from Dr. Joel Gelernter and Dr. Henry R. Kranzler for providing clinical samples for this study.
References
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, et al. (1998) Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry 55: 967-972.
- Kendler KS, Jacobson KC, Prescott CA, Neale MC (2003) Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry 160: 687-695.
- Edenberg HJ, Foroud T (2006) The genetics of alcoholism: identifying specific genes through family studies. Addict Biol 11: 386-396.
- Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, et al. (2006) Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet 15: 807-819.
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, et al. (2006) Association of the kappa-opioid system with alcohol dependence. Mol Psychiatry 11: 1016-1024.
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J (2008) The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry 13: 531-543.
- Corbett AD, Henderson G, McKnight AT, Paterson SJ (2006) 75 years of opioid research: the exciting but vain quest for the Holy Grail. Br J Pharmacol 147 Suppl 1: S153-162.
- Bodnar RJ, Klein GE (2006) Endogenous opiates and behavior: 2005. Peptides 27: 3391-3478.
- Wise RA, Bozarth MA (1987) A psychomotor stimulant theory of addiction. Psychol Rev 94: 469-492.
- Koob GF, Bloom FE (1988) Cellular and molecular mechanisms of drug dependence. Science 242: 715-723.
- Ritz MC, Lamb RJ, Goldberg SR, Kuhar MJ (1987) Cocaine receptors on dopamine transporters are related to self-administration of cocaine. Science 237: 1219-1223.
- Kuhar MJ (1998) Recent biochemical studies of the dopamine transporter--a CNS drug target. Life Sci 62: 1573-1575.
- Brodie MS, Pesold C, Appel SB (1999) Ethanol directly excites dopaminergic ventral tegmental area reward neurons. Alcohol Clin Exp Res 23: 1848-1852.
- Spanagel R, Herz A, Shippenberg TS (1992) Opposing tonically active endogenous opioid systems modulate the mesolimbic dopaminergic pathway. Proc Natl Acad Sci U S A 89: 2046-2050.
- Shippenberg TS, LeFevour A, Heidbreder C (1996) kappa-Opioid receptor agonists prevent sensitization to the conditioned rewarding effects of cocaine. J Pharmacol Exp Ther 276: 545-554.
- Schoffelmeer AN, Hogenboom F, Mulder AH (1987) Inhibition of dopamine-sensitive adenylate cyclase by opioids: possible involvement of physically associated mu- and delta-opioid receptors. Naunyn Schmiedebergs Arch Pharmacol 335: 278-284.
- Rogers H, Henderson G (1990) Activation of mu- and delta-opioid receptors present on the same nerve terminals depresses transmitter release in the mouse hypogastric ganglion. Br J Pharmacol 101: 505-512.
- Gray AC, Coupar IM, White PJ (2006) Comparison of opioid receptor distributions in the rat central nervous system. Life Sci 79: 674-685.
- Vaught JL, Takemori AE (1979) Differential effects of leucine and methionine enkephalin on morphine-induced analgesia, acute tolerance and dependence. J Pharmacol Exp Ther 208: 86-90.
- Heyman JS, Jiang Q, Rothman RB, Mosberg HI, Porreca F (1989) Modulation of mu-mediated antinociception by delta agonists: characterization with antagonists. Eur J Pharmacol 169: 43-52.
- Gomes I, Jordan BA, Gupta A, Trapaidze N, Nagy V, et al. (2000) Heterodimerization of mu and delta opioid receptors: A role in opiate synergy. J Neurosci 20: RC110.
- Pan ZZ (1998) mu-Opposing actions of the kappa-opioid receptor. Trends Pharmacol Sci 19: 94-98.
- Funada M, Suzuki T, Narita M, Misawa M, Nagase H (1993) Blockade of morphine reward through the activation of kappa-opioid receptors in mice. Neuropharmacology 32: 1315-1323.
- Jordan BA, Devi LA (1999) G-protein-coupled receptor heterodimerization modulates receptor function. Nature 399: 697-700.
- Li Z, Zheng T, Califano A, Floratos A (2007) Pattern-based mining strategy to detect multi-locus association and gene x environment interaction. BMC Proc 1 Suppl 1: S16.
- Wilcox MA, Li Z, Tapper W, Browning S, Curtin K, et al. (2007) Genetic association with rheumatoid arthritis-Genetic Analysis Workshop 15: summary of contributions from Group 2. Genet Epidemiol 31 Suppl 1: S12-21.
- Pfeiffer A, Brantl V, Herz A, Emrich HM (1986) Psychotomimesis mediated by kappa opiate receptors. Science 233: 774-776.
- Longmate JA (2001) Complexity and power in case-control association studies. Am J Hum Genet 68: 1229-1237.
- Lazzeroni LC, Lange K (1998) A conditional inference framework for extending the transmission/disequilibrium test. Hum Hered 48: 67-81.
- McIntyre LM, Martin ER, Simonsen KL, Kaplan NL (2000) Circumventing multiple testing: a multilocus Monte Carlo approach to testing for association. Genet Epidemiol 19: 18-29.
Relevant Topics
- Addiction Recovery
- Alcohol Addiction Treatment
- Alcohol Rehabilitation
- Amphetamine Addiction
- Amphetamine-Related Disorders
- Cocaine Addiction
- Cocaine-Related Disorders
- Computer Addiction Research
- Drug Addiction Treatment
- Drug Rehabilitation
- Facts About Alcoholism
- Food Addiction Research
- Heroin Addiction Treatment
- Holistic Addiction Treatment
- Hospital-Addiction Syndrome
- Morphine Addiction
- Munchausen Syndrome
- Neonatal Abstinence Syndrome
- Nutritional Suitability
- Opioid-Related Disorders
- Relapse prevention
- Substance-Related Disorders
Recommended Journals
Article Tools
Article Usage
- Total views: 14287
- [From(publication date):
specialissue-2013 - Dec 22, 2024] - Breakdown by view type
- HTML page views : 9781
- PDF downloads : 4506