Research Article Open Access
A Short Interfering RNA (siRNA) Molecular Beacon for the Detection of Mycobacterial Infection
Remo George1, Norman Bolus1, Shawn Williams2, Joseph Garner1, Kathy Nugent1 and M. Tino Unlap3*1Departments of Clinical and Diagnostic Sciences, University of Alabama at Birmingham, USA
2The Vision Science Research Center, University of Alabama at Birmingham, USA
3Departments of Clinical and Diagnostic Sciences* and Biochemistry and Molecular Genetics, University of Alabama at Birmingham, AL 35294, USA
- Corresponding Author:
- M. Tino Unlap
Department of Clinical and Diagnostic Sciences
SHPB476, University of Alabama at Birmingham, AL 35294, USA
Tel: 205-934-7382
E-mail: unlap@uab.edu
Received date: June 30, 2012; Accepted date: August 18, 2012; Published date: August 21, 2012
Citation: George R, Bolus N, Williams S, Garner J, Nugent K, et al. (2012) A Short Interfering RNA (siRNA) Molecular Beacon for the Detection of Mycobacterial Infection. J Biotechnol Biomater 2:147 doi:10.4172/2155-952X.1000147
Copyright: © 2012 George R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
In latent TB, the ability of Mycobacterium tuberculosis to invade and survive within macrophages of the pulmonary granuloma is attributed to protein products of mammalian cell entry (mce4) operon genes (A-F). These are cholesterol transporters which facilitate the transport of host lipids into the mycobacterium allowing long term survival during chronic infection. Currently, there are no rapid and reliable tests for the detection of latent TB. Therefore, because there is a lack of reliable and efficient tests for the diagnosis of latent TB, we tested the hypothesis that mycobacterial infection can be detected using mce4 siRNA molecular beacons against mce4 mRNAs. Because our work showed that the mce4A gene of the mce4 operon conferred infectivity to host E. coli, a siRNA molecular beacon was designed against a region of the mce4A mRNA that is highly homologous in Mycobacterium tuberculosis and Mycobacterium smegmatis. This molecular beacon has a hairpin structure with a stem, 5 nucleotides on either end that are complementary to each other, and a loop which contains 20 nucleotides that are complementary to a region of the target mRNA. Conjugated to the 5’ and 3’ ends of the molecular beacon are the fluorophore TYE 665 and quencher Iowa Black RQ-SP. In the absence of the target mRNA the hairpin structure will predominate and fluorescence will be quenched while in the presence of the target mRNA fluorescence will be induced. Our study shows that the siRNA molecular beacon detects its target in M. smegmatis and in macrophages infected with M. smegmatis and offers a potential test for detection of mycobacterial infection.
Keywords
Latent TB; Molecular beacon; Mce4; Mycobacteria; Sirna
Introduction
The persistence of latent Tuberculosis (TB) continues to be a significant problem in the world today, more so for third world countries [1]. Latent TB is characterized by pulmonary granulomas which allow Mycobacterium tuberculosis (Mtb) to survive for years without detection [2]. Most of those with latent TB infection are unaware of their condition until their infection becomes acute which occurs in over 10% of latent TB cases [3]. As the world becomes easier to travel, especially through modern modes of transportation, latent TB has become every one’s problem even for highly industrialized countries like the U.S. M. tuberculosis survival in granulomas is possible through its ability to synthesize a mammalian cell entry protein, mce4 [4]. This protein is encoded on the mce4 operon that consists of five genes designated mce4A-F. The products of these genes are cholesterol transporters that help transport lipids from the host macrophage into the mycobacterium and allows the mycobacterium to survive for years during chronic infections [5]. Currently there are no reliable, rapid tests for the detection of latent TB and because this is asymptomatic it is difficult to treat [6]. Therefore, because there is lack of reliable and efficient tests for the diagnosis of latent TB, this study was conducted in order to design a siRNA molecular beacon against one of the mammalian cell entry protein genes that could be used for the detection of mycobacterial infection in macrophages.
More than 80% of tuberculosis (TB) cases in the United States are from reactivation of latent TB infection [6]. The hallmark of pulmonary TB is the granulomas harboring the bacterial infection along with their draining lymph nodes. Each granuloma has a necrotic core in the center that provides nutritional source for the persisting Mtb bacteria and is surrounded by concentric layers of macrophages, epitheloid cells, multinucleated Langhans giant cells, and lymphocytes [3]. These TB lesions are surrounded by highly vascularized tissue [7] which enables the targeting of latent Mtb with systemically delivered drugs. The treatment for active or latent TB infection consists of an extended course of antibiotics spanning many months using drugs like isoniazid or rifampicin, which generally carries poor patient compliance rates [6]. Unfortunately, all available tests today can only strongly suggest the presence of latent TB but cannot confirm it. This is due to low specificity for chest x-ray, low sensitivity for tuberculin skin test in BCG vaccinated individuals, or low sensitivity for interferon gamma release assay in children, immune-compromised persons, and the elderly. Currently, there are no direct imaging methods for locating TB bacteria in the body, which can be critical when trying to identify high risk individuals for prophylactic regimen as well as for identifying extrapulmonary TB sites in HIV co-infected patients. Developing a direct M. tuberculosis imaging screening tool for the asymptomatic population is going to be vital in the fight against tuberculosis.
Studies showed that mce operons are widely distributed throughout the genus Mycobacterium, and M. smegmatis, a non-pathogenic mycobacterial species that shares many features with M. tuberculosis, possesses a homolog of mce4 [8,9]. Owing to its lower biosafety level restrictions and the presence of an identical MCE4 cholesterol transport system,M. smegmatis (Ms) provides a safe mycobacterial model for preliminary studies.
Because over 90% of the people infected with Mtb will have latent TB (LTB) infection and 10% will eventually go on to have full-blown active TB at a later stage in their life [10], prompt identification and treatment of individuals with LTB is important for the effective control of this disease. Currently, there are no tests available to directly detect the presence of Mtb in an affected individual and assessment of latent TB infection involves an imperfect approach of measuring the host immune response to mycobacterial infection [6]. This testing deficiency can be especially critical when trying to identify high risk individuals for prophylactic regiment as well as identifying extrapulmonary TB sites in HIV co-infected patients [11]. Developing a direct Mtb imaging-screening tool for the asymptomatic population along with novel treatment strategies is vital to our fight against tuberculosis.
Molecular beacons (MB) are hairpin shaped single stranded nucleic acid probes that fluoresce only upon hybridization with its target molecule. They have a stem-loop structure, with a fluorophore and a quencher attached to opposite ends. The on/off signals produced by the florophore/quencher pair depends on the conformational state of the MB. In the absence of mce4 mRNA, the stem, which consists of four to seven base pairs, brings the quencher nearby the fluorophore and turns the fluorescence off with high quenching efficiency via Fluorescence Resonance Energy Transfer (FRET). In the presence of the target mce4 mRNA, the loop region of the molecular beacon hybridizes with the target mRNA and opens up the hairpin structure into a linear structure, thus causing separation of the fluorophore and the quencher which results in increase in fluorescence and identification of the target mycobacterium [12].
Latent TB is a silent epidemic that threatens the development and fundamental progress of many societies across the globe. In order to counteract this epidemic, rapid detection and effective treatment for TB is critical. To that end, we will test the hypothesis that Mycobacterial smegmatis infection can be detected in macrophages using a siRNA molecular beacon against one of the mce4 operon genes. The findings of these studies will demonstrate the utility of detecting mycobacterial infection using siRNA molecular beacons which can then be easily adapted to detecting infection in animal models and eventually in humans.
Methods
Mammalian cell culture
Breast cancer MCF7 cells and U937 human monocytic leukemia cells were purchased from the ATCC and were maintained in HQ-DMEM or RPMI medium supplemented with penicillin and streptomycin at 1 unit/ml and enriched with 10% (v/v) fetal bovine serum and 2 mM L-glutamine. Cells were routinely passaged every other day. Cells were cultured in flasks at 37°C for propagation and in 12 well plates with glass cover slips for differentiation (U937 cells) and infection. U937 cell differentiation was achieved as described previously [13]. Briefly, this was done by incubating 1 × 106 cells with complete RPMI medium supplemented with 4 nm PMA. After 2 days of treatment with PMA (differentiation) nonadherent cells were aspirated, coverslips were removed and adherent cells were counted using a haemocytometer with viable cells identified by trypan blue dye exclusion.
Bacterial strains and culture conditions
M. smegmatis mc2 155 was purchased from ATCC. E. coli Top10, obtained from Invitrogen, was used as the host bacteria for cloning experiments.M. smegmatis mc2 155 was grown in Middlebrook 7H9 broth (Difco) with 0.05% Tween 80 and supplemented with OADC (oleic acid, albumin, glucose, catalase supplement). E. coli cultures were grown on Lennox L (LB) broth and antibiotics were added as appropriate (50 μg/ml ampicillin for E. coli harboring pTrcHis2- TOPO). All cultures were incubated at 37°C and shaken at 190 rpm.
Genomic DNA isolation from M. smegmatis
Genomic DNA fromM. smegmatis was isolated from a 100 ml culture ofM. smegmatis which was grown to an OD600 of 1.6. Approximately 1 x 109 cells were harvested by centrifugation (12,000 g for 1 min) and genomic DNA was extracted using the Axyprep Bacterial Genomic DNA Miniprep Kit (Axygen Biosciences, CA) according to the manufacturer’s protocol. The purified genomic DNA was used for the PCR amplification of specific mce4 operon genes.
PCR Amplification
PCR was performed using genomic DNA (50 ng) fromM. smegmatis and 100 ng of gene specific forward and reverse primers in 25 μL containing GoTaq®; Master Mix (Promega). Genomic DNA fromM. smegmatis was used along with five sets of primers to amplify a 1.2 Kb mce4A fragment (5′-GAGGAGCCATGGATGTCGAACGGAAACGCCAAA- 3′/5′-GGAAGGAAGC TTGAAGTCGTCCCTTTCCGCGAA-3′), a 1.1Kb mce4B fragment (5′-GGAAAGCGATCGTTCTAGATGCACCGCGACAGG-3′/5′- GCCATTCTCCGAGCACCTCCC-3′), a 1.0 Kb mce4C fragment (5′-TGGCGACTTCGGCCTCACGAT- 3′ / 5′- CGGAGAATGGCTAGTCTGCGC- 3′), a 1.4 Kb mce4D fragment (5′-TCCGCCCGCACCTCCGGGAGC- 3′/5′-GTCGCCATGACACATTCGAAT-3′) and a 1.7 Kb mce4F fragment (5′-CCGTAGATGATCGACCGGCTG-3′ / 5′- AGCCTGCCTTGGATCCAGCAT- 3′).
PCR was carried out using gene specific conditions. Ten microliters of each PCR product was transferred to a separate tube and mixed with 2 μL of 6x DNA loading dye followed by electrophoresis on a 1% agarose gel at 70 v for one hour along with 1 Kb DNA marker. Samples containing the correct fragment sizes were used for subsequent cloning reactions.
Cloning of mce4 Genes
Ligation of each PCR fragment into the vector was achieved by mixing an aliquot (2 μL) of the PCR sample with 1μL of pTrcHis2- TOPO and 2 μL of sterile deionized water (SDW) followed by incubation at room temperature for 5 minutes, transformation and plating on agar plates containing 50 μg/ml of ampicillin and incubated overnight at 37°C. Four colonies were selected and grown over night in L-broth (Difco) containing 50 μg/ml ampicillin. Colonies were screened by taking 500 μl aliquot of each overnight culture and centrifuging at 13,000 xg for 5 min, lysed in 100 μl of 1X STE (100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA) by vigorous vortexing followed by mixing with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), and then vortexed and centrifuged at 13,000 × g for 2 minutes. A 20 μl aliquot of the aqueous layer (top) was mixed with 4 μl of 6x DNA loading dye and resolved by agarose gel. The clones which showed a 4.4 Kb plus the correct sizes of inserts were used for plasmid isolation using the SNAP Midiprep Kit (Invitrogen) according to the manufacturer’s instructions. In order to determine the orientation of each insert, two plasmids representing each gene were sequenced in both directions by SeqWright (SeqWright, Inc). E. coli containing each of the mce4 genes (E.coli-4A-F) in the correct orientation and the vector alone (E.coli- TOPO) were used for functional assays.
Invasion assay time-course
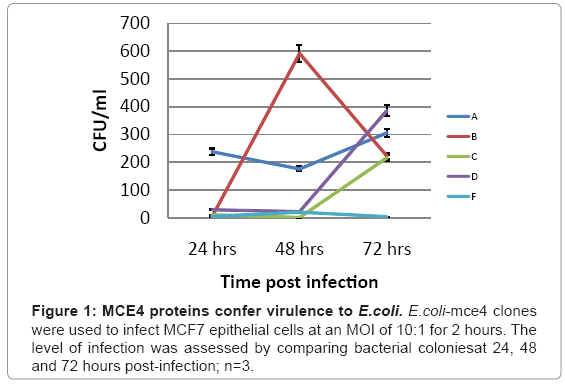
In order to determine the virulence that each gene conferred to the host E. coli, MCF7 cells were seeded at 2.5 × 105 cells per well in 12-well plates and incubated for 24 hrs. MCF7 cells were incubated in fresh medium (HyQ DMEM supplemented with 5% FCS and 2 mM L-glutamine) at 37°C for 30 min and E.coli, E.coli-TOPO or E.coli-4A-F were added to each well at a multiplicity of infection (MOI) of 10:1 and incubated at 37°C for 3 hours. Cells were washed 3 times with HyQ DMEM media which contained 5% fetal bovine serum (FBS), 1% penn/ strep and 100 μg/ml kanamycin to remove extracellular bacteria and lysed after 24, 48 and 72 hrs. For lysis, cells were incubated for 10 min in 500 μl of lysis buffer (0.1% Triton X-100 in PBS, pH 7.4) and the lysate was plated on LB agar plates containing ampicillin (100 μg/ml) and incubated at 37°C overnight. Recombinant E. coli colonies were counted and the numbers of E. coli that survived at 24, 48 and 72 hrs post infection was plotted versus time.
mce4A siRNA molecular beacon
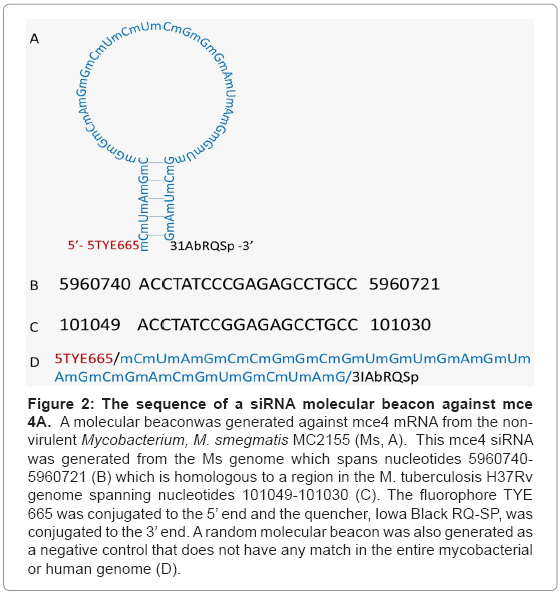
Our functional assays demonstrated that the mce4A gene conferred virulence to the host E. coli which appeared early and was sustained for the entire infection period. Therefore, a molecular beacon siRNA was designed against the mce4A gene. This mce4 siRNA was generated from theM. smegmatis genome which spans nucleotides 5960740- 5960721 and is homologous to a region in the M. tuberculosis H37Rv genome spanning nucleotides 101049-101030. The fluorophore TYE 665 was conjugated to the 5’ end and the quencher, Iowa Black RQ-SP, was conjugated to the 3’ end.
Confocal Imaging
An overnight culture ofM. smegmatis MC2 155 was used to inoculate 7H9 broth at 1/50th volume and grown to an A600 of 0.3. Mce4 siRNA molecular beacon or random oligonucleotide molecular beacon was added at 10 μM to the M. smegmatis, incubated at 37°C for 5 hrs and imaged using a confocal microscope.
Detection of M. smegmatis in differentiated U937 cells
To determine the ability of the mce4A siRNA molecular beacon to detect mycobacterial infection in macrophages, differentiated U937 cells were either not infected or infected at a multiplicity of infection (MOI) of 10:1 bacteria (E.coli-4A) per macrophage in a well and incubated at 37°C for 3 hours to allow for phagocytosis to occur. Each well was washed 3 times with RPMI 1640 media which contained 10% fetal bovine serum (FBS), 1% penn/strep and 100 μg/ml kanamycin to remove extracellular bacteria. Fresh RPMI media, 3 ml, containing the appropriate antibiotic was added to each well containing the infected or non-infected cells and incubated at 37°C in the presence or absence of 1 or 10 μM mce4A siRNA molecular beacon for 3 hours. Fluorescence measurements were carried out using a microplate reader, GloMaxTMat peak emission wavelength of 665 nm.
Results
Cloning of mce4 operon genes
Using gene specific forward and reverse primers, each mce4 gene was PCR amplified from M. smegmatis. Using the pTrcHis2-TOPO cloning system, cloning of the five constituent genes of the mce4 operon ofM. smegmatis was accomplished in a timely manner. However, since the genes were amplified through PCR using gene specific primers cloning of each gene into the vector was non directional. Therefore, two positive clones were selected for sequencing to ascertain orientation. The reverse primer for each gene excluded the stop codon in order to express the c-myc and 6x His tags with each protein product and only clones containing the gene and c-myc and 6x His tags that were in frame were selected for use in subsequent experiments.
mce4 operon genes confer virulence to E. coli
Which of the five mce4 operon genes is primarily responsible for latent TB infection is not currently known. Therefore, the ability of each of the five mce4 operon genes to confer virulence to the host E.coli was determined by comparing the number of colonies that grew on agar plates following lysis of MCF7 cells which were infected with E. coli expressing the vector (E.coli-TOPO)or each of each of the five mce4 operon genes (E. coli-4A-F). The results showed that while mce4A, B, C, and D genes confer virulence to the host E. coli, mce4A showed virulence that appeared early and was sustained for 24, 48 and 72 hr post infection (Figure 1).
The design of a mce4A siRNA molecular beacon
Because mce4A conferred virulence to the host E.coli which appeared early and was sustained for the entire 72 hr period, a siRNA molecular beacon was designed against this gene (Figure 2). This siRNA molecular beacon consists of 20 nucleotides which are complementary to a region of the mce4A mRNA and 8 nucleotides on the 5’ end and another 8 nucleotides on the 3’end. The 8 nucleotides on either end of the structure are complementary to each other such that they form a double stranded stem for the molecular beacon. Also conjugated to the 5’ and 3’ ends of the molecular beacon were the fluorophore TYE 665 and the quencher Iowa Black RQ-SP, respectively. The rationale in the design was that in the absence of its target mRNA, the mce4A molecular beacon will be in the form of a hair loop structure and fluorescence will be quenched. In the presence of the target mRNA, however, the molecular beacon will open up and bind with the 20 complementary nucleotides to the mRNA. This will position the fluorophore and the quencher apart such that fluorescence will not be quenched.
Figure 2: The sequence of a siRNA molecular beacon against mce 4A. A molecular beaconwas generated against mce4 mRNA from the nonvirulent Mycobacterium, M. smegmatis MC2155 (Ms, A). This mce4 siRNA was generated from the Ms genome which spans nucleotides 5960740- 5960721 (B) which is homologous to a region in the M. tuberculosis H37Rv genome spanning nucleotides 101049-101030 (C). The fluorophore TYE 665 was conjugated to the 5’ end and the quencher, Iowa Black RQ-SP, was conjugated to the 3’ end. A random molecular beacon was also generated as a negative control that does not have any match in the entire mycobacterial or human genome (D).
The mce4A siRNA molecular beacon interacts with its target in M. smegmatis
Once the mce4A siRNA was designed, it was necessary to demonstrate that it would interact with its mce4A mRNA target in M.smegmatis. Mce4 siRNA molecular beacon or random oligonucleotide molecular beacon was added at 10 μM to the M. smegmatis, incubated at 37°C for 5 hrs and imaged using a confocal microscope. Confocal microscopy imaging demonstrated that the siRNA but not random oligonucleotide molecular beacon interacted with its target mRNA inM. smegmatis (Figure 3).
Figure 3: mce4A siRNA molecular beacon interacts with its target in M. smegmatis. An overnight culture of M. smegmatis MC2155 was used to inoculate 7H9 broth and grown to an A600 of 0.3. Mce4 siRNA molecular beacon or random oligonucleotide molecular beacon was added at 10μM to the M. smegmatis, incubated at 37°C for 5hrs and imaged using a confocal microscope.
The mce4A siRNA molecular beacon interacts with its target in M. smegmatis
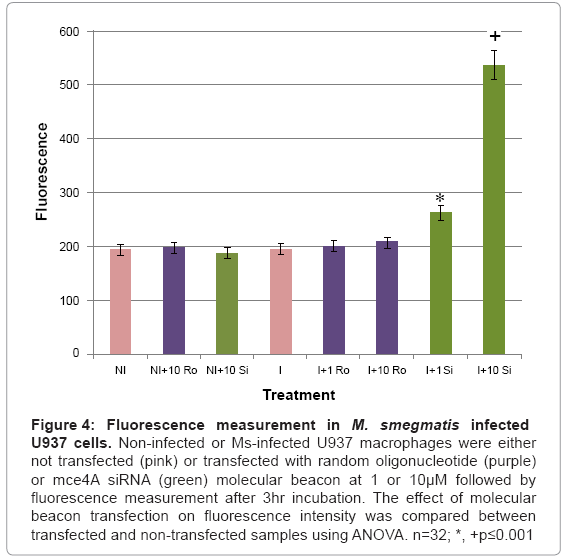
The designed mce4A siRNA molecular beacon was used for the detection of M. smegmatis infection in infected U937 cells. Infected and noninfected U937 cells were transfected with 1 or 10 μM of the siRNA or a random oligonucleotide molecular beacon, incubated overnight followed by fluorescence measurement using a GloMax®- Multi microplate reader. Our results (Figure 4) demonstrated that baseline fluorescence was observed in noninfected cells not transfected or transfected with 10 μM of the siRNA or random oligonucleotide molecular beacon and in infected cells which were not transfected or transfected with 1 or 10 μM of the random oligonucleotide molecular beacon. Cells infected withM. smegmatis and transfected with the siRNA molecular beacon showed significant levels of fluorescence which were above baseline levels. Comparison of fluorescence between baseline levels and fluorescence levels in infected cells transfected with 1 or 10 μM of the mce4A siRNA molecular beacon using Analysis of Variance (ANOVA) indicated significance at p ≤ 0.001.
Figure 4: Fluorescence measurement in M. smegmatis infected U937 cells. Non-infected or Ms-infected U937 macrophages were either not transfected (pink) or transfected with random oligonucleotide (purple) or mce4A siRNA (green) molecular beacon at 1 or 10μM followed by fluorescence measurement after 3hr incubation. The effect of molecular beacon transfection on fluorescence intensity was compared between transfected and non-transfected samples using ANOVA. n=32; *, +p≤0.001
Discussion
The use of molecular beacons for in vivo detection of mRNA is gaining popularity. These small hairpin structures are stable in cellular environments and bind to their target mRNAs with a high degree of specificity. They can be easily modified by the addition of dyes and quenchers in order to enhance their utility in detection protocols. Molecular beacons have been used in a number of areas including detection of specific mRNA targets in vivo [14] and in cervical living cells including cancer cells [15,16], identification of allelic variance for drug resistance [17], and detection of organisms for diagnostic tests [17-20].
Small interfering RNA (siRNA) technology has been used to inhibit transcription for hepatitis G virus [21], influenza virus [22], picorna virus [23], and trypanosma brucei [24]. The potential combination of the two technologies, molecular beacon and siRNA, in the health care industry is tremendous. Molecular beacon siRNA have been used for knocking down the expression of telomerase mRNA in human breast cancer cells [25], BMP4 mRNA in hedge hog signaling [16], and aromatase mRNA in breast cancer cells [25]. Thus, siRNA can be designed in the form of molecular beacons in order to be used in detection and therapeutics.
This study was necessitated by three things, 1) latent TB continues to be a problem not only for third world countries but also for industrialized countries like the U.S., 2) because the mycobacterium growth rate is so slow and specific diagnostic tests for latent TB are currently not available which naturally leads to infection, 3) there is a lack of specific and efficient treatment for latent TB. To assist in the effort to detect and treat latent TB, this study was conducted in order to test the hypothesis that a molecular beacon siRNA designed against the mce4 operon, which has been shown to be responsible for latent TB infection [4,26], especially mce4A [26,27], could be used for detection and attenuation of mycobacterial infection in macrophages. This hypothesis was tested in MCF7 breast cancer cells and in differentiated U937 cells.
In order to design a molecular beacon siRNA, it was necessary to determine which of the mce4 operon genes confer the highest degree of virulence to host E. coli. Because the M. tuberculosis has a slow growth rate and there is high degree of homology between mce4 operons of M. tuberculosis and other mycobacteria [8], the mce4 operon of the rapid growingM. smegmatis was selected. Using gene specific primers with the reverse primer for each set excluding the termination codon, mce4A-F were PCR amplified, cloned into the prokaryotic expression vector pTrcHis2-TOPO and stably expressed in E.coli. Western blot assays using c-myc and 6x His monoclonoal antibodies showed that the proteins were expressed in host E.coli. Invasion assays in MCF7 breast cancer cells (Figure 1) showed that mce4A-F conferred virulence to the host E.coli. However mce4A conferred virulence which was high from the beginning and was sustained during the entire invasion period (72 hr). Therefore mce4A was selected to be the target for a molecular beacon antisense RNA (Figure 2).
The mce4A molecular beacon antisense RNA was designed to contain a double stranded stem which consists of nucleotides that are complementary to each other to form a 5-base pair double stranded stem. The loop consists of 20 nucleotides that are complementary to a region of the target mce4A mRNA and conjugated to the 5’ and 3’ ends of this molecular beacon are the fluorophore TYE 665 and quencher Iowa Black RQ-SP respectively. The double stranded stem facilitates cytosolic localization of the siRNA [28] and in the presence of the target mce4A mRNA the siRNA molecular beacon will bind and fluorescence intensity will increase, since the fluorophore and the quencher are separated, and degradation of the target mRNA will be induced.
This molecular beacon design combines both detection and therapeutic [16,25,29]. The rationale is that in the absence of the target mce4A mRNA the molecular beacon remains in its hairpin form while in the presence of its target mRNA the 20 nucleotide loop will compete with the 5 nucleotide stem for hybridization to their targets, the loop to the mce4A mRNA and the stem to its complementary pair on the opposite end of the molecular beacon. The hybridization of the loop, based on the number of nucleotides (20 versus 5), to its target will be greater than that of the strands for the stem. Hybridization of the loop to the mce4A mRNA will separate the fluorophore from the quencher which will induce fluorescence (detection) and degradation (therapeutic) of the mRNA. Because the mycobacterium utilizes the product of mce4A for survival on cholesterol for carbon and energy source [26,27,30,31], degradation of the mce4A mRNA will lead to reduced survival. This study tested the ability of the mce4A siRNA to detect its target mce4A mRNA inM. smegmatis and in macrophages infected with M. smegmatis. The results show that the molecular beacon siRNA detects its target inM. smegmatis and in macrophages infected with M. smegmatis. Thus, a molecular beacon can be designed against one of the mce4 operon genes inM. smegmatis that facilitates the detection of mycobacterial infection in macrophages. Current tests are being carried out to test the ability of this siRNA molecular beacon to not only detect but also attenuate mycobacterial infection in macrophages.
These studies were set up as proof of concept for the detection of mycobacterial infection in macrophages. Since these studies indicate that a mce4A siRNA molecular beacon could be used for the detection of mycobacterial infection in macrophages, future studies would be conducted to explore the utility of this technology for the detection and eradication of mycobacterial infection in animal models which could be adapted to detection and eradication in humans using a nebulizer which lends itself to patenting. This will not only speed up the detection but enhance the eradication of LTB. It is unlikely that this technology will lead to reactivation of LTB since detection and eradication are simultaneous. In order to make this technology accessible to third world countries, less expensive but more sensitive fluorophores and quenchers must to be explored.
Acknowledgements
We would like to thank the Nuclear Medicine Technology and Clinical Laboratory Science Programs for the support they provided for this study. We would also like to thank the Vision Science Research Center for the use of the High Resolution Imaging Facility and Shawn Williams for his assistance with the confocal microscopy work. Remo George is a graduate student in the department of Biochemistry and Molecular Biology and is supported by a grant from the American Society of Radiological Technology Research Foundation (Award# 2008942)
References
- WHO (2010) Tuberculosis global facts 2010/2011. Cent Eur J Public Health 18: 197.
- Barry CE 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, et al. (2009) The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol 7: 845-855.
- Mariano M (1995) The experimental granuloma. A hypothesis to explain the persistence of the lesion. Rev Inst Med Trop Sao Paulo 37: 161-176.
- Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW (1993) Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 261: 1454-1457.
- Pandey AK, Sassetti CM (2008) Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A 105: 4376-4380.
- Horsburgh CR Jr, Rubin EJ (2011) Clinical practice. Latent tuberculosis infection in the United States. N Engl J Med 364: 1441-1448.
- Ulrichs T, Kosmiadi GA, Jorg S, Pradl L, Titukhina M, et al. (2005) Differential organization of the local immune response in patients with active cavitary tuberculosis or with nonprogressive tuberculoma. J Infect Dis 192: 89-97.
- Haile Y, Caugant DA, Bjune G, Wiker HG. (2002) Mycobacterium tuberculosis mammalian cell entry operon (mce) homologs in Mycobacterium other than tuberculosis (MOTT). FEMS Immunol Med Microbiol 33: 125-132.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403-410.
- Kumar V, Abbas KA, Fausto N, Mitchell R Robbins basic pathology. (8thedn). Philadelphia, PA: Saunders/Elsevier.
- Chakraborty MS, Chakraborty A (2000) Tuberculosis and HIV illness. J Indian Med Assoc 98: 103-106, 109.
- Kim Y, Sohn D, Tan W (2008) Molecular beacons in biomedical detection and clinical diagnosis. Int J Clin Exp Pathol 1: 105-116.
- Adunyah SE, Unlap TM, Franklin CC, Kraft AS (1992) Induction of differentiation and c-jun expression in human leukemic cells by okadaic acid, an inhibitor of protein phosphatases. J Cell Physiol 151: 415-426
- Bratu DP, Catrina IE, Marras SA (2011) Tiny molecular beacons for in vivo mRNA detection. Methods Mol Biol 714: 141-157.
- Xue Y, An R, Zhang D, Zhao J, Wang X, et al. (2011) Detection of survivin expression in cervical cancer cells using molecular beacon imaging: new strategy for the diagnosis of cervical cancer. Eur J Obstet Gynecol Reprod Biol 159: 204-208.
- Rhee WJ, Santangelo PJ, Jo H, Bao G (2008) Target accessibility and signal specificity in live-cell detection of BMP-4 mRNA using molecular beacons. Nucleic Acids Res 36: e30.
- Balashov SV, Gardiner R, Park S, Perlin DS (2005) Rapid, high-throughput, multiplex, real-time PCR for identification of mutations in the cyp51A gene of Aspergillus fumigatus that confer resistance to itraconazole. J Clin Microbiol 43: 214-222.
- Morandi L, Ferrari D, Lombardo C, Pession A, Tallini G (2007) Monitoring HCV RNA viral load by locked nucleic acid molecular beacons real time PCR. J Virol Methods 140: 148-154.
- Santangelo P, Nitin N, LaConte L, Woolums A, Bao G (2006) Live-cell characterization and analysis of a clinical isolate of bovine respiratory syncytial virus, using molecular beacons. J Virol 80: 682-688.
- Warren DK, Liao RS, Merz LR, Eveland M, Dunne WM Jr (2004) Detection of methicillin-resistant Staphylococcus aureus directly from nasal swab specimens by a real-time PCR assay. J Clin Microbiol 42: 5578-5581.
- Cao M, Ren H, Zhao P, Pan W, Zhao L, et al. (2005) Small interfering RNA-mediated inhibition of hepatitis G virus gene expression in human hepatoma cell Huh-7. Sci China C Life Sci 48: 61-69.
- Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, et al. (2003) RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci U S A 100: 2718-2723.
- Lim T, Yuan J, Zhang HM, Sall A, Liu Z, et al. (2008) Antisense DNA and RNA agents against picornaviruses. Front Biosci 13: 4707-4725.
- Shi H, Tschudi C, Ullu E (2007) Depletion of newly synthesized Argonaute1 impairs the RNAi response in Trypanosoma brucei. RNA 13: 1132-1139.
- Zhou C, Mao Y, Sugimoto Y, Zhang Y, Kanthamneni N, et al. (2011) SPANosomes as Delivery Vehicles for Small Interfering RNA (siRNA). Mol Pharm 9: 201-210.
- Saini NK, Sharma M, Chandolia A, Pasricha R, Brahmachari V, et al. (2008) Characterization of Mce4A protein of Mycobacterium tuberculosis: role in invasion and survival. BMC Microbiol 8: 200.
- Xu G, Li Y, Yang J, zhou X, Yin X, et al. (2007) Effect of recombinant Mce4A protein of Mycobacterium bovis on expression of TNF-alpha, iNOS, IL-6, and IL-12 in bovine alveolar macrophages. Mol Cell Biochem 302: 1-7.
- Chen AK, Davydenko O, Behlke MA, Tsourkas A (2010) Ratiometric bimolecular beacons for the sensitive detection of RNA in single living cells. Nucleic Acids Res 38: e148.
- Chang E, Zhu MQ, Drezek R (2007) Novel siRNA-based molecular beacons for dual imaging and therapy. Biotechnol J 2: 422-425.
- Miner MD, Chang JC, Pandey AK, Sassetti CM, Sherman DR (2009) Role of cholesterol in Mycobacterium tuberculosis infection. Indian J Exp Biol 47: 407-411.
- Senaratne RH, Sidders B, Sequeira P, Saunders G, Dunphy K, et al. (2008) Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J Med Microbiol 57: 164-170.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 15183
- [From(publication date):
August-2012 - Nov 25, 2025] - Breakdown by view type
- HTML page views : 10398
- PDF downloads : 4785