Make the best use of Scientific Research and information from our 700+ peer reviewed, Open Access Journals that operates with the help of 50,000+ Editorial Board Members and esteemed reviewers and 1000+ Scientific associations in Medical, Clinical, Pharmaceutical, Engineering, Technology and Management Fields.
Meet Inspiring Speakers and Experts at our 3000+ Global Conferenceseries Events with over 600+ Conferences, 1200+ Symposiums and 1200+ Workshops on Medical, Pharma, Engineering, Science, Technology and Business
Research Article Open Access
A Rapid Approach for Detecting Seven Genetically Modified Canola Events using Optical Thin-Film Biosensor Chips
| Sulan Bai1*, Liang Xu1, Xiaomin Guo1, Yaochuan Zhang2, Tingting Deng3, Wensheng Huang3, Ying Chen3 and Yikun He1 | |
| 1Key laboratory of Genetics and Biotechnology, College of Life Sciences, Capital Normal University, Beijing 100048, P.R. China | |
| 2Beijing Vocational College of Agriculture, Beijing 102442, P.R. China | |
| 3Institute of Food Safety, Chinese Academy of Inspection and Quarantine, Beijing 100123, P.R. China | |
| Corresponding Author : | Sulan Bai Key laboratory of Genetics and Biotechnology College of Life Sciences, Capital Normal University Beijing 100048, P.R. China Tel: 86-10-68981191 Fax: 86-10-68981191 E-mail:sulanb@sina.com |
| Received : November 10, 2011; Accepted March 14, 2012; Published March 16, 2012 | |
| Citation: Bai S, Xu L, Guo X, Zhang Y, Deng T, et al. (2012) A Rapid Approach for Detecting Seven Genetically Modified Canola Events using Optical Thin-Film Biosensor Chips. J Biochip Tissue chip 2:101. doi:10.4172/2153-0777.1000101 | |
| Copyright: © 2012 Bai S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited | |
Visit for more related articles at Journal of Bioengineering and Bioelectronics
| Keywords |
| Genetically modified canola; Event-specific; Optical thin-film biosensor chip; Microarray |
| Introduction |
| With the increasing development of genetic engineering, the industrialization of transgenic crops has become an irresistible trend. In recent decades, hundreds of genetically modified (GM) plants have been approved for commercialization worldwide [1]. For instance, in 2004, seven GM canola events (Ms1×Rf1, Ms1×Rf2, Ms8×Rf3, GT73, T45, Oxy-235, and Topas 19/2) from the United States and Canada were approved for commercialization [2,3]. Clearly, GM plants can bring enormous economic and social benefits. However, there are also environmental and food safety risks which make it necessary to implement mandatory labeling for foods derived from GM plants [1]. Consequently, the development of reliable methods for detecting and identifying GM plants has become increasingly important for government regulators, international trade organizations [4]. |
| Currently Polymerase Chain Reaction (PCR) technology is the most popular technique for the detection and identification of transgenic crops [5,6,7]. PCR technology has proven to be a highly sensitive and specific method for GM plant identification via amplification of event-specific genes. This method, however, is problematic when analyzing several similar targets or detecting a particular target which may be present in several GM crops [8]. Multiplex PCR strategy is an accepted mean to detect multiple targets in a single assay [9]. Common problems of multiplex PCR are decreased sensitivity and increased nonspecific amplification products [10]. Real-time PCR is frequently used for quantitative and qualitative analysis of species-specific genes [11]. However, it is expensive and prone to generating false positive signals [12]. |
| One possible means for overcoming these limitations is a microarray- based technology. The basic idea involves hybridization of labeled PCR products to a DNA microarray [13]. However, the expensive equipment needed for scanning the arrays may limits its general usage in many laboratories. For rapid, efficient, accurate, and inexpensive detection, we developed a method for the detection of seven GM canola events (Ms1×Rf1, Ms1×Rf2, Ms8×Rf3, GT73, T45, Oxy-235, and Topas 19/2) on the surface of optical thin-film biosensor chips. The detection results can be visualized by the naked eye. This approach not only takes advantage of microarrays, but also obviates the need for preparing the expensive equipments [14]. This technique has been verified as a useful method for detecting seven GM canola lines rapidly and economically. |
| Materials and Methods |
| Preparation of samples |
| Genetically modified canola events (Ms1×Rf1, Ms1×Rf2, Ms8×Rf3, GT73, T45, Oxy-235 and Topas 19/2) were friendly supplied by Chinese Academy of inspection and quarantine, Beijing, PR China. |
| DNA Extraction |
| Plant genomic DNA extraction was conducted as CTAB method [15]. The DNA concentration was measured using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). DNA samples were diluted to100 ng/uL for further evaulation. |
| Primer and probe synthesis |
| In this study, we want to detect seven GM canola events (Ms1×Rf1, Ms1×Rf2, Ms8×Rf3, GT73, T45, Oxy-235, and Topas 19/2) including three rapeseed hybrids. So we need to design oligonucleotide primers and probes for nine events (Ms1, Rf1, Ms1, Rf2, Ms8, Rf3, GT73, T45, Oxy-235 and Topas 19/2) respectively. Primers and probe were designed according to the unique and specific integration junction sequences between the inserted foreign genes and the recipient canola genomes. A pair of primers, one from the recipient genome DNA, another from the integrated genome, was originated from published papers and shown in Table 1. The genetic elements located in the seven GM canola lines are shown in Figure 1. The 5’end of the reverse primers for each gene has a biotin modification. There is an aldehyde modification at the 5’end of the probes and it is followed by ten deoxyadenosine residues. The details about probes and primers are shown in Table 1. |
| PCR amplification |
| PCR reactions were conducted according to the method described by Su et al [15]. Briefly, the primary PCR was carried out in a volume of 25 μl mixing containing 1×PCR buffer (50 mM KCl, 10 mM Tris- HCl,pH 8.3, 1.5 mM MgCl2), dNTP (0.1 mmol/L), primers (0.2 mmol/L each), genomic DNA library (100ng) and rTaq (1 U; TaKaRa, Japan) in a nested approach. Amplifications were executed as follows: heating at 94°C for 5 min for pre-denaturation, 35 cycles of 30 s at 94°C, 55°C for 30 s, and 72°C for 30s, followed by the last step of 10 min at 72°C. The PCR amplicons were then analyzed by 2% agarose gel. |
| Optical thin-film biosensor chips |
| Biosensor chips work as follows: when the white light irradiates on the optical biosensor chip, it initially reflects a golden color, and, the color changes with the thickness of the optical layer. It had been verified that such changes could even be visualized by the naked eye [14]. The biosensors were prepared following the procedure described by Jenison et al. [16] and Bai et al. [14]. |
| Standard assay procedures |
| To detect nucleic acid target analytes, the aldehyde-labeled probes (250 nL or 40nL per spot) were spotted on to biosensor chips by manual pipet or a robotic pipetting device (Biodot AD3200). Add PCR amplification targets onto chips two hours after spotting probes. The PCR amplification targets (100 fmol each in 100 μL of reaction buffer) were denatured at 95°C for 5 min first and then hybridized on the chip for 15 min at 50°C in hybridization buffer [5×standard saline citrate (SSC), 5 mg/ml acid-treated caseine (ATC)]. After three washes with 0.1×SSC, the chips were incubated with an anti-biotin IgG-horseradish peroxidase (HRP) conjugate (Jackson Immuno Research; 3:1000 dilution from a 1 mg/ml stock in a buffer containing 5×SSC: 5 mg/mL, ATC: 10% glycerol) for 12 min in hybridization buffer. After three washes in 0.1×SSC, 100 μL of tetramethylbenzidine (TMB, BioFx Laboratories, Owings Mills, MD) was added and incubated for 10 min at room temperature. The chips could be scored by eye without any help when they were dried after washed in water. The chips can also be imaged with a digital camera shown in this study. |
| Results and Discussion |
| Assay principle |
| The event-specific detection strategy based on the unique and specific integration junction sequences between the inserted foreign genes and the recipient canola genome has been utilized because of its high specificity (Figure 1). For the hybrid canola, there are two foreign genes followed by two specific sequences. Thus, there is a total of nine specific sequences used for detection in this study. The thin-film biosensor chips used for detection of GM canola described here depends on hybridization of biotinylated PCR fragments incubated with probes on optical surfaces [17]. For each gene, a primer pair and a probe were synthesized. The 5’ end of the probes was aldehyde-labeled so they could be covalently attached to the chip surface. In addition, the probes had 10dA as spacers, followed by about a 40 bp nucleotide sequence corresponding to the sequence between the forward and reverse primers of the specific sequences [14]. Samples containing target nucleic acids were hybridized to the chip surface. To facilitate detection, the reverse primer was labeled directly with biotin at the 5’ end (Table 1). Following 10 min of hybridization between capture probes and PCR fragments, the immobilized target sequence was treated with an antibiotin IgG-HRP conjugate and a precipitable HRP substrate. The mass contributed by the precipitate alters the path length of light reflected from the chip surface resulting in a distinguishable color change from gold to blue which could be read either by the naked eye or by a simple digital camera [18]. |
| Defining sensitivity and specificity of thin-film biosensor chips |
| To estimate the limits of the methods in this study, we randomly chose two event-specific probes, GT73 and T45, and diluted them to 0.01, 0.1, 1, and 10 μM before manually spotting them on the biosensor chip surface at a volume of 250 nL per spot (Figure 2, left panel). GT73 event-specific gene PCR amplification was performed in the range of 0, 0.1, 1, 10, and 100 fmol. The chips were then hybridized with PCR products in 100 μL total reaction volume. Only spots containing GT73 event-specific probes were detected (Figure 2, right panel), thus con-firming the specificity of this assay. The test showed that the signal intensity declined with diminution of the concentrations of probe and target DNA. Moreover, the signals were detected when the probe was only at a concentration of 0.01 μM in the presence of 100 fmol of PCR products. According to the results of the test, we chose 1μM as the optimal probe density to be spotted on biosensor chips. |
| Amplification of nine target gene fragments by PCR |
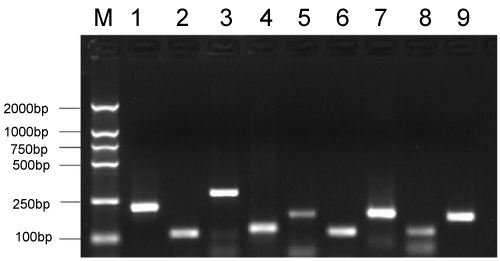
| The PCR products for seven GM canola events (Ms1×Rf1, Ms1×Rf2, Ms8×Rf3, GT73, T45, Oxy-235, and Topas 19/2) including the nine events (Ms1, Rf1, Ms1, Rf2, Ms8, Rf3, GT73, T45, Oxy235, and Topas 19/2) are shown in (Figure 3). The sizes of amplified fragments were in line with expectations (Table 1). The results showed that the specificity of the PCR amplification of the target genes was quite accurate. |
| Chip design and detection of target genes using optical thin film biosensor chips |
| The chips spotted by robotic pipetting (40 nL per spot) were used to detect seven GM canola lines as shown in Figure 4. M refers to the positive control biotin-dA20 which always exhibited a signal if the chip detection system worked. In this detection system, all other genes were negative controls for the target gene. The results showed that specific signals were detected for all seven genetically modified canola lines (Figure 4). For example, T45 event-specific targets amplified (Figure 4,lane1) were used for hybridization to a biochip and the assay resulted in one set of colored dots (Figure 4, lower panel, 1). The other eight gene targets showed their own specific sets of colored dots (Figure 4). For the transgenic rapeseed hybrids (Ms1×Rf1, Ms1×Rf2 and Ms8×Rf3), every hybrid shows two sets of signal dots. For example, Ms1×Rf1 shows signals at position seven and eight (data not show). From the detection results, we can see that no false-positive signals were observed. This study indicates that one designed chip can be readily used to detect the presence of these GM canola lines. |
| Conclusions |
| Event-specific assays for identification of seven GM canola lines were established based on optical thin-film biosensor chips. Through analyses of the specificity, repeatability and accuracy, the methods established in this paper were found to be easy, sensitive, accurate, and suitable for the event-specific qualitative detection of seven GM canola lines. In addition, this technique does not require any specific instruments for data scanning, and may be readily utilized in a variety of laboratory settings. |
| Acknowledgements |
| This work was supported by Beijing Municipal Education Commission (KM200800005004), Natural Science Foundation of China (No.30771094), Beijing Natural Science Foundation (5082003, 5112006). |
| References |
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
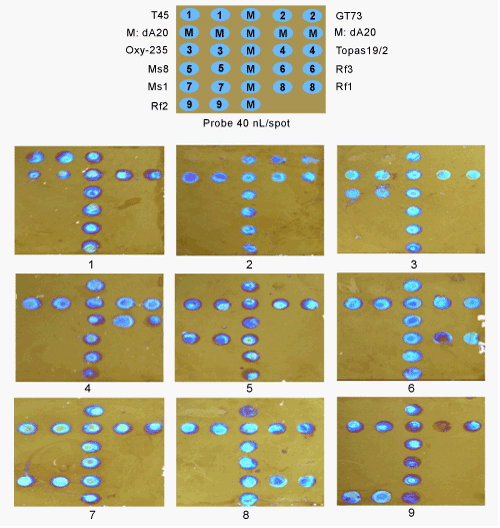 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Post your comment
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14324
- [From(publication date):
April-2012 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 9859
- PDF downloads : 4465
