Research Article Open Access
A pH Responsive Ternary Gene Carrier based on Branched- Poly(ethylenimine) and Poly(2-ethyl-2-oxazoline)-block- Poly(methacrylic acid)
Che-Ping Lin1, Yueh-Che Sung1 and Ging-Ho Hsiue1,2*1Department of Chemical Engineering, National Tsing Hua University, 300 Taiwan, R.O.C
2Department of Chemical Engineering, Chung Yuan Christian University, 200 Taiwan, R.O.C
- Corresponding Author:
- Prof. G.H. Hsiue
Department of Chemical Engineering, National Tsing Hua University
101, Section 2 Kuang Fu Road, Hsinchu, 300 Taiwan, ROC
E-mail: ghhsiue@mx.nthu.edu.tw
Received date: June 10, 2011; Accepted date: July 07, 2011; Published date: July 09, 2011
Citation: Lin C, Sung Y, Hsiue G (2011) A pH Responsive Ternary Gene Carrier based on Branched - Poly(ethylenimine) and Poly(2-ethyl-2-oxazoline)-block- Poly(methacrylic acid). J Biotechnol Biomaterial 1:109. doi:10.4172/2155-952X.1000109
Copyright: © 2011 Lin C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
This study demonstrates new ternary polyion gene carriers, based on the PH.-responsive diblock copolymer poly(2-ethyl-oxazoline)-block-poly(methacrylic acid) (PEOz-b-PMAA), and the branched-poly (ethylenimine) (B-PEI). This study complexes the plasmid DNA with B-PEI and further with PEOz-b-PMAA to obtain ternary polyplexes (DNA/B-PEI/PEOz-b-PMAA). PMAA were partially dissociated under neutral PH. with a negative charge, to attach to the positive charge surface of the B-PEI/DNA polyplex. The ternary polyplexes will also desorb and return to the original pre-poly complex to help gene release after cell uptake due to PMMA become neutral charge under an acid environment in endosome. The ternary polyplexes showed suitable mean particle size, low cytotoxicity, and acceptable transfection at PH 7.4 because of shielding of B-PEI by PEOz-b-PMAA. TEM morphology showed that the stable core-shell structure of ternary polyplexes at PH. 7.4 collapsed and released plasmid at PH. 5. Observations of cell uptake of the B-PEI/DNA polyplex and ternary polyplexes by CLSM revealed that ternary polyplexes started to accumulate after 3 h incubation and accumulated significantly after 6 h. In conclusion, the ternary polyplex improves cytotoxicity of the single B-PEI/DNA polyplex, and presents a PH.-responsive behavior to enhance gene escape from the polyplex. The ternary polyplex constitutes a useful approach for gene carrier design.
Keywords
PH-Responsive; Ternary gene carrier; Poly (2-Ethyl-Oxazoline); Poly (Methacrylic Acid)
Introduction
Non-viral vectors, such as cationic polymers, peptides, and liposomes, have attracted interest because of their advantages compared to viral systems. For example, research suggests that non-viral vectors are safer, with more flexible structures and chemical properties [1-3]. Among the various types of polymer gene carriers, branched polyethylenimine (B-PEI) is the most effective vector for delivering genes because the BPEI/DNA polyplexes exhibit great transgene expression in vitro and in vivo [4,5]. The high transfection efficiency of B-PEI is because of its high buffering capacity, which disrupts the endosomal membrane, and releases the polyplex into the cytoplasm [6]. The intrinsic high cytotoxicity of B-PEI limits its application in vivo. Excessive positive charge on the cationic polymer carriers’ surface is toxic to cells and tissues [7], and adversely influences biodistribution of the complexes in vivo [8]. Poly (2-ethyl-2-oxazoline) (PEOz) is a water-soluble polyelectrolyte, with low toxicity and high hydrophilicity similar to poly (ethylene glycol) (PEG) [9] and it is approved by the FDA as a biocompatibility polymer. The charge density in the linear polyethylenimine (LPEI) in our past research strongly affected cell viability, and the PEOz-coupled LPEI reduced cationic charge density and cytotoxicity [10].
Studies in recent years have examined the multilayer gene carriers made by “layer by layer self-assembly” [11,12]. Trubetskoy used the poly-L-lysine (PLL) and succinylated PLL (SPLL) as polycations and polyanions to form the DNA/PLL/SPLL complex with a charge stoichiometry close to 1:1:1. The zeta potential of the previously mentioned complexes became negative, indicating effective surface recharging [13]. Koyama reported a new ternary gene carrier structure by preparing the carriers from DNA polycation as the core structure (binary polyplex), and covering them with a negative charge of PEG-carbocylic acid. This ternary polyplex showed a steady property instead of the binary polyplex appearing as an aggregated state in BSA [14]. As the cell uptake carried the polyplex into the endosome, the carbocylic acids protonated and lost hydrophilicy. The structure of the hydrophilic acids fused with the endosome membrane [15], to enhance escape from the endosome.
This study proposes a special ternary complex vector for the encapsulating gene, considering the advantage of the multilayer gene carrier. The study complexes the plasmid DNA with the branchedpoly( ethylenimine) (B-PEI), to form the pre-polyplex B-PEI/DNA, and with the diblock copolymer poly (2-ethyl-oxazoline)-blockpoly( methacrylic acid) (PEOz-b-PMAA) to obtain ternary polyplexes (DNA/B-PEI/PEOz-b-PMAA) (Figure-1). This study partly dissociates the PMAA with a negative charge under neutral PH, to attach to the positive charge surface of the B-PEI/DNA polyplex. The ternary polyplexes (DNA/B-PEI/PEOz-b-PMAA) desorbs and returns to the original prepoly complex to help gene release after cell uptake because PMMA has a neutral charge under an acid environment. The gene easily escapes from the polyplex, given the previously mentioned phenomenon. This work evaluates the physicochemical and biological characteristics of the ternary complex micelles.
Materials and Methods
2-ethyl-2-oxazoline (EOz) (Aldrich) and methyl p-toluenesulfo nate (Aldrich) were purified by vacuum distillation over CaH2. Tetrahydrofuran (THF) (TEDIA), chloroform (TEDIA), and acetonitrile (ACN) (Fisher Chemical) were dried over CaH2 and distilled under dry nitrogen. 4-(dimethylamino) pyridinium-4-toluene-sulfonate (DPTS) was obtained by mixing with hydrated p-toluenesulfonic acid (Fluka) and toluene (Aldrich) at 85°C and then with 4-dimethylaminopyridine (Aldrich) at 40°C. Methacrylic acid (MAA) (Aldrich) was purified by vacuum distilled. N,N’-dicyclohexylcarbodimide (DCC), Triton X-100, 4’,6-diamidino-2-phenylindole (DAPI) and uranyl acetate were obtained from Aldrich. Dimethyl sulfide (DMSO), N,N’-dimethylformamide (DMF) and dichloromethane (DCM) were obtained from TEDIA. 4,4’-azobis(4-cyanovaleric acid) (ABPCA), branched-polyethylenimine (B-PEI) 25 kDa were obtained from Fluka. Ethidium bromide (EtBr) and 3-(4,5-dimethylthiazol-2-yl)- 2,5-diphe-nyl tetrazolium bromide (MTT) were purchased from ICN Biomedicals, Inc. pUHC 13-3 encoding for luciferase as a reporter gene was received as a gift from Professor Jia-Ling Yang from National Tsing Hua University, Taiwan. The lucifierase kit #E1531 was obtained from Promega. Cyanin 5.5 was obtained from GE Healthcare. LysoTracker yellow HCK-123 was purchased from Invitrogen.
Synthesis of poly (2-ethyl-2-oxazoline) (PEOz) and poly (2-ethyl-2-oxazoline)-4, 4’-Azobis (4-cyanovaleric acid) (PEOz-ABCPA)
Poly (2-ethyl-2-oxazoline) (PEOz) was synthesized by cationic ring-opening polymerization of 2-ethyl-2-oxazoline using methyl ptoluenesulfonate as an initiator. A solution of methyl p-toluenesulfonate (305.04 mg, 1.64 mmol; 182.28 mg, 0.98 mmol and 120.9 mg, 0.65 mmol) and 2-ethyl-2-oxazoline (10 ml, 99.19 mmol) in dry acetonitrile (30 ml) was heated to 110°C and stirred for 20 h in a nitrogen atmosphere. The living PEOz polymer chains were terminated by adding KOH-methanol solution at 0°C. The polymer solution was filtered through the silica gel and purified by precipitation in diethyl ether. After drying, the experiment obtained a white powdery product, PEOz. PEOz was coupled with ABCPA with DCC at 0°C. DPTS was used as a catalyst. The side products, including PEOz-ABPCPA, PEOz, and other impurities were removed by a stirred ultrafiltration cell with suitable dialysis membranes. The solvent was removed by freeze-drying and obtaining (PEOz)2-ABCPA. The composition was analyzed by 1H NMR and GPC.
Synthesis of poly (2-ethyl-2-oxazoline)-poly (methacrylic acid), (PEOz-b-PMAA)
The diblock copolymer PEOz-b-PMAA was synthesized by free radical polymerization with the macroinitiator (PEOz) 2-ABCPA. The experiment dissolved (PEOz) 2-ABCPA in dichloromethane and heated it until 80°C, before adding MAA and stirring for 18 h. The product was purified by dialysis for 2 d using cutoff dialysis tubing with a molecular weight of 6000, and dried in a vacuum, obtaining the PEOz-b- PMAA product. The composition was analyzed by 1H NMR.
Preparation of B-PEI/DNA polyplex and PEOz-PMAA/BPEI/ DNA ternary polyplex
Polyplex preparation included diluting the required amount of polymer and 1 μg of luciferase encoded plasmid pUHC-13-3 in 100 μl of distilled water and gently mixing. The polyplex formulation was incubated at room temperature for 30 m before using. After preparing the B-PEI/DNA polyplex, the ternary polyplex was created by mixing the B-PEI/DNA solution and the PEOz-b-PMAA solution, and incubated for 30 m before using.
Measurement of particle size and zeta potential
To measure the particle size and zeta potential, various formulations of polyplexes were prepared. Dynamic light scattering was used to decide the particle size of polyplex particles in distilled water at 25°C (Zetasizer 3000HS, Malvern Instruments, Worcestershire, U.K). Laser doppler micro-electrophoresis in the capillary electrophoresis cell of the Zetasizer 3000HS at 25°C determined the zeta potential of polyplexes prepared in distilled water. The mean values for these measurements were calculated from the data obtained in ten runs.
Cell culture and MTT assay
Human cervix carcinoma HeLa cells (1 x 104 cells/well) were cultured onto a plate with ninety-six wells in DMEM media supplemented with 10% FBS in a humid atmosphere of 5% CO2 at 37°C for 24 h. Then the growth medium was replaced with 200 μl medium that contained the desired amount of polymers and allowed to incubate for 72 h. The cytotoxicity of each sample was determined by measuring the cell viability using a tetrazolium dye (MTT) assay. 100 μl of the medium containing 10 μl of MTT PBS solution (5 mg/ml) was added. After incubation for 2 h, the formazan crystals were dissolved in 100 μl of DMSO/ EtOH solution. The absorbance of each well was measured using a microplate reader (Stat Fax 2100, Awareness, Palm City, FL, USA) at a test wavelength of 570 nm and a reference wavelength of 630 nm.
In vitro transfection and total protein assay
To evaluate the transfection potential of polyplexes, human cervix carcinoma HeLa cells (2 x 105 cells/well) were incubated in DMEM media supplemented with 10% FBS and plated in six-well plates in a humid atmosphere of 5% CO2 at 37°C for 24 h. The growth medium (2 mL) contained DNA or polyplexes that contained 5 μg of plasmid incubated at 37°C. After incubation for 48 h, the cells were washed with cold PBS and then lysed using 400 μL of the 1X cell lysis buffer. The cell lysate was then transferred into Eppendorf tubes and centrifuged for 15 s at 12,000 rpm. Then luciferase activity was measured in relative light units (RLU), using a ninety-six-well plate luminometer (Wallac 1420 Multilabel Counter, Perkin-Elmer, USA) and a luciferase assay kit (Promega, Tokyo, Japan). The total amount of protein was measured at 595 nm using a protein assay.
Morphology of the polyplex
The polyplex morphology prepared as described above was observed by transmission electron microscopy (TEM, Hitachi, H-7500, Tokyo, Japan). To prepare the TEM sample, 10 μl of polyplex solution was placed onto a copper grid coated with carbon, tapped with a filter paper to remove water, and air-dried for 5 m. Then, the grid was negatively stained with 2 wt% uranyl acetate solution for 30 s, tapped with a filter paper to remove water, and vacuum-dried for 24 h.
Cellular uptake of polyplexes
The amino end group of B-PEI was used to label the Cy5.5 dye to observe the cellular uptake behavior of polyplexes by a confocal laserscanning microscope (CLSM). B-PEI was mixed with Cy5.5 NHS ester in DMSO at room temperature for 24 h. The product was purified by dialysis for 2 d in a dark bottle. Polyplex accumulated in HeLa cells was localized using LSM5 PASCAL CLSM (Carl Zeiss, USA). The HeLa cells were seeded on cover-slides for 24 h and were then treated with free DNA or polyplexes. After specific time, the cells were washed twice with PBS, and then LysoTracker was added to the culture medium without FBS and incubated for 20 m. Then the samples were stored at 4°C refrigerator for 16 h, removed from the formaldehyde solution, and we added DAPI water solution (10μg/ml) for 5 m.
Result and Discussions
Polymer synthesis
Scheme 1 shows the synthesis of PEOz-b-PMAA. PEOz was coupled with ABCPA in DCC to form the macroinitiator (PEOz)2-ABCPA. The catalyst, DPTS, prepared by equal mole PTSA and DMAP, improves efficiency of the esterification reaction. The process removed the side products, which are PEOz-ABPCPA, PEOz, and other impurities, by stirring with the ultrafiltration cell with suitable dialysis membranes. The single peak in GPC indicated obtaining the pure macroinitiator (PEOz)2-ABCPA. The 1H-NMR spectrum of (PEOz)2-ABCPA dissolved in CDCl3 is: δ0.96 (s,-CH2CH3 from the ethyl group of PEOz); δ2.042~2.253 (m,-CH2CH3 from the ethyl group of PEOz); δ0.96 (s,– NCH2CH2 from the main chain of PEOz); δ1.554~1.612 (m, CH3 from ABCPA); δ2.865~3.105 (m, CH2CH2 from ABCPA).The previously mentioned macroinitiator (PEOz)2-ABCPA synthesized the diblock copolymer PEOz-b-PMAA. The PEOz-b-PMAA product produced strong hydrogen bonding in water solution during the dialysis process. A weak base environment (PH=5) should separate the unreacted MAA and PEOz-b-PMAA, destroying the hydrogen bonding between PEOzb- PMAA polymers. The 1H-NMR spectrum of the PEOz-b-PMAA dissolved in CDCl3 is: δ0.96 (s,-CH2CH3- from the ethyl group of PEOz); δ2.042~2.253 (m,-CH2CH3 from the ethyl group of PEOz); δ0.96 (s,– NCH2CH2 of the PEOz main chain); δ1.554~1.612 (m, CH3 from ABCPA); δ2.865~3.105 (m, CH2CH2 from ABCPA); δ0.91~1.02 (m,-CH3 from the methyl group of PMAA ); δ1.68~1.90 (m,-CH2 of the PMAA main chain). The experiment calculated the molecule weight of PMMA by comparing the integral peak area of the methylen groups of PMMA (δ1.68~1.90) to that of the ethylene groups of PEOz from the 1H-NMR spectrum.
Table 1 presents the compositions of the PEOz-b-PMAA. The average molecular weight and polydispersity index of PEOz determined by GPC are 5300, 10500, 13900 and 1.15, 1.06, 1.11. The code “ExMy” represents x, y for the molecular weight of the PEOz and PMMA. For example, the code E6M2 means PEOz-b-PMAA with a PEOz molecular weight of approximately 6000 (corresponds to 5300), and a PMMA molecular weight of approximately 2000 (corresponds to 2100). This study used the previously mentioned copolymers to follow the experiments.
| Polymer | Code | Mn of PEOza | PDIa | Mn of PMAAb | Total Mn |
|---|---|---|---|---|---|
| PEOz6k-PMAA2k | E6M2 | 5300 | 1.15 | 2100 | 7400 |
| PEOz6k-PMAA4k | E6M4 | 5300 | 1.15 | 4100 | 9400 |
| PEOz10k-PMAA2K | E10M2 | 10500 | 1.06 | 1200 | 11700 |
| PEOz10k-PMAA4K | E10M4 | 10500 | 1.06 | 1400 | 11900 |
| PEOz15k-PMAA2k | E15M2 | 13900 | 1.11 | 1300 | 15200 |
aEvaluate by GPC at 40℃, 1 ml/s, solvent: NMP
bEvaluate by 1H-NMR 500MHzdd
Table 1: Compositions and characterization of diblock copolymer PEOz-b-PMAA.
Measurement of particle size and zeta potential
Laser light scattering estimated the particle size and zeta potential of the polyplexes. The polyplexes prepared in an aqueous solution of physiological salt (150 mM NaCl) or in distilled water measured the polyplex size to explain whether the ionic strength of the media influences polyplex stability.
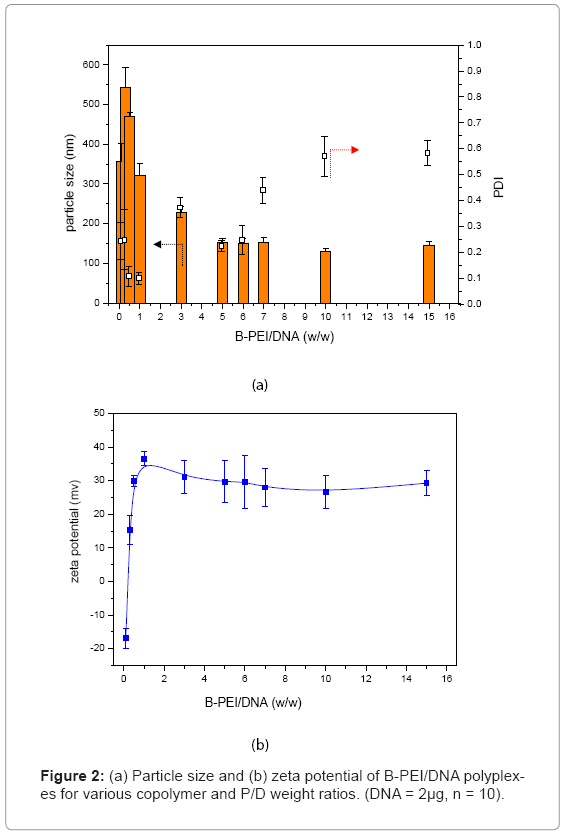
Prepolycomplex B-PEI/DNA: To form prepolycomplex B-PEI/ DNA, plasmid DNA was complexed with B-PEI. Figure 2 shows the particle size and zeta potential of the B-PEI/DNA polyplex in distilled water. The B-PEI/DNA polyplex formed large particles (particles size were greater than 300 nm) as the polymer/DNA weight ratio (μg/μg) dropped below 1. When the B-PEI/DNA weight ratio was above 5, the mean diameter of particle sizes was around 150 nm, and the tendency kept constant. The PDI of the B-PEI/DNA polyplex grew after the weight ratio went above 5. The zeta potential of the polyplex expressed a similar tendency. Initially, the zeta potential indicated a negative charge as with a B-PEI/DNA weight ratio below 1. With increasing weight ratio of B-PEI/DNA, the zeta potential became positive, and the value maintained an almost constant value of 30 mV with a weight ratio above 5. The previously mentioned results indicated that the B-PEI/ DNA polyplex formed a most stable conformation with a weight ratio around 5. The following experiments adopted the B-PEI/DNA weight ratio of 5, and selected the B-PEI/DNA weight ratio of 1?3 as contrast forms. The code “B-PEI mX” represents m for the B-PEI/DNA weight ratio (μg/μg). For example, the code B-PEI 5X means the B-PEI/DNA weight ratio equals 5, as used below.
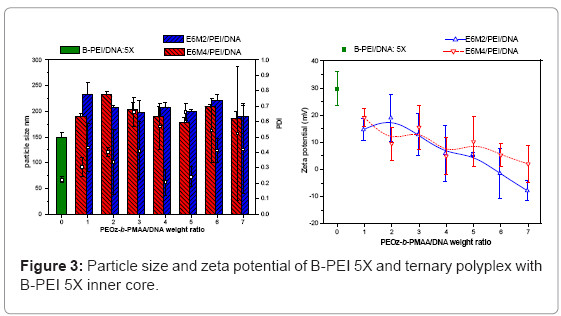
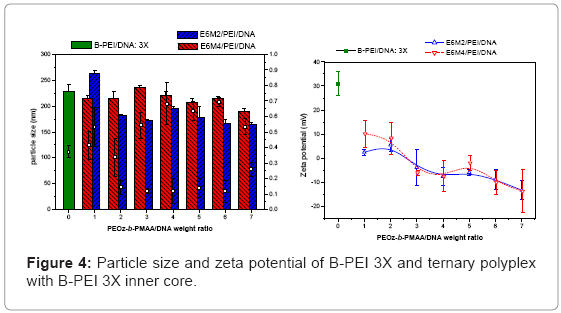
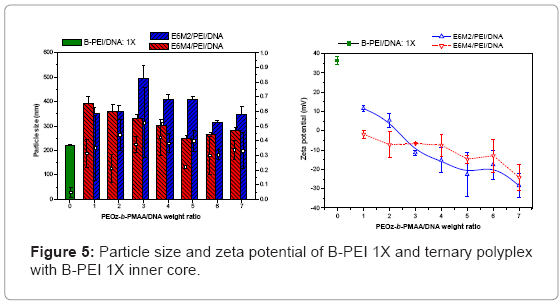
Ternary polyplexes: This study prepared five compositions of PEOz-b-PMAA (E6M2, E6M4, E10M2, E10M4, and E15M2) to form ternary polyplexes by covering PEOz-b-PMAA polymers on the B-PEI/ DNA core structure. The ternary polyplexes with a longer hydrophilic segment PEOz from PEOz-b-PMAA (E10M2, E10M4, and E15M2) showed higher particle size (above 200 nm) and zeta potential (around 30 mV). Increased positive charges and large size may be due to the higher hydrophilic force from the longer PEOz segment, exposing the positively charged PEI parts to the outer polyplex. The section below discusses the two kinds of PEOz-b-PMAA with a short PEOz segment (that is, E6M2 and E6M4). Figure 3 shows the results of particle size and zeta potential for the inner core, B-PEI 5X, and the outer shell, PEOz-b-PMAA (E6M2 and E6M4), with various weight ratios based on DNA in distilled water. Comparing it with the pre-poly complex B-PEI/ DNA, the particle size increased slightly with a mean diameter from 150 nm to about 200 nm, but zeta potential decreased from 30 mV to about 10 mV. The results indicated that as the ternary polyplex weight ratio of PEOz-b-PMAA increased, the particle size and zeta potential decreased. This change may be due to the increasing neutralization effect between negative charges from PMMA and positive charges from B-PEI. Figure 4 and Figure 5 show similar results for B-PEI 3X and B-PEI 1X, but zeta potential decreased to about -20 mV. It may be due to the ternary polyplexes having more negative charges from PMMA at the same weight ratio conditions.
The B-PEI parts are well-set inside the polyplex attracted by DNA, and the neutral PEOz parts are expected to come outside to provide less charge to the polyplex surface. The hydrophilic and neutral segment, PEOz, creates an optimal particle size and zeta potential to the polyplex for cell entry. The incoming negative charges from PMAA cover the positive charges of B-PEI/DNA. The greater the weight ratio of B-PEI/ DNA is, the more PEOz-b-PMAA is needed to neutralize the B-PEI/ DNA positive charge. In other words, the smaller the weight ratios of B-PEI/DNA are, the greater the negative trend for the zeta potential of the ternary polyplex. (Results of gel retardation assay are shown in supplementary information).
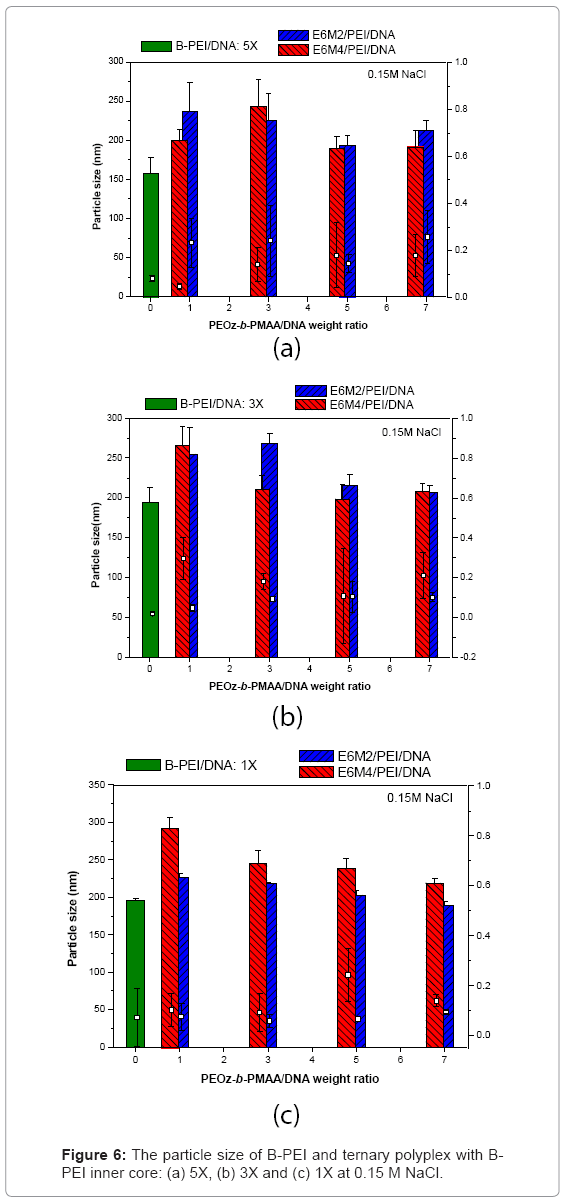
Polyplexes in salt-containing media: Figure 6 shows the results of ternary polyplexes prepared in an aqueous solution of 150 mM NaCl to discuss the stability of ternary polyplexes. The ternary polyplexes did not aggregate in salt-containing media. The results indicated that the neutral BPEI and LPEI polyplexes were prone to aggregate, because of the lack of electrostatic repulsion. The DNA/B-PEI/PEOz-b-PMAA ternary polyplexes were stable because they possessed a core-shell structure with a hydrophobic B-PEI/DNA core and a hydrophilic PEOz shell that prevented the polyplexes from aggregating.
In vitro transfection and total protein assay
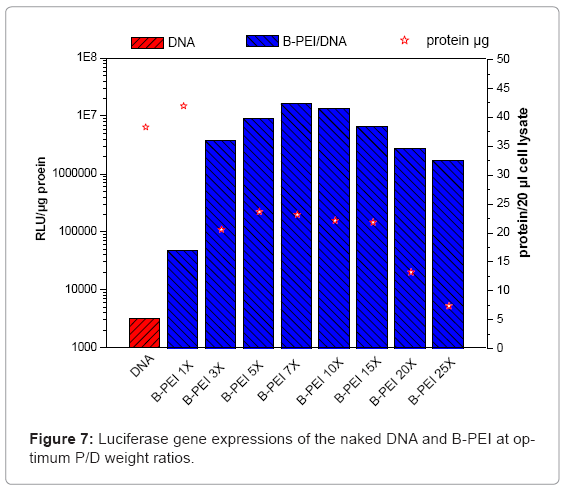
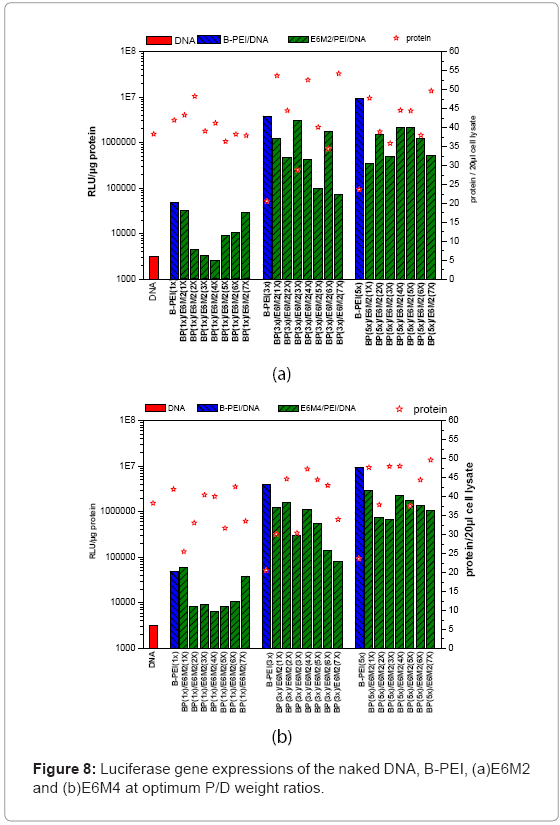
This study evaluated in vitro transfection efficiencies of naked DNA, B-PEI/DNA polyplexes, and DNA/B-PEI/PEOz-b-PMAA (E6M2, E6M4) ternary polyplexes under various P/D ratios on the HeLa cell. The luciferase assay for the luciferase encoding plasmid determines transfection efficiency. This work used total protein production, an indirect measure of induced toxicity, to normalize the RLU. Figures 7 and Figures 8 present luciferase gene expressions of the naked DNA, B-PEI, E6M2, and E6M4 at optimum P/D weight ratios. The transfection using polyplexes resulted in greater efficiency compared to when using the naked DNA. The naked DNA with negative charges repelled the cell as it approached negatively charged cell membranes, resulting in poor transfection efficiency. The B-PEI polyplex transfection efficiency was approximately proportional to the weight ratio of B-PEI/DNA; the more B-PEI, the better the transfection result. Although the B-PEI polyplex has the highest gene expression, it has the lowest protein production, indicating the highest toxicity. A higher amount of B-PEI may kill the cell or retard physiological operation. Transfection decreased with a B-PEI/DNA weight ratio of 7.
This work compared the transfections of ternary polyplexes with B-PEI/DNA polyplexes. The ternary polyplex prepared from B-PEI 3X showed a decreasing tendency with increasing PEOz-b-PMAA. However, the ternary polyplex formed from B-PEI 5X showed better transition efficiency than the ternary polyplex formed from B-PEI 3X. The ternary polyplex formed from B-PEI 5X did not decline, but remained at the same level. The above phenomena match the results of zeta potential, the surplus negative charges from PMMA of ternary polyplex prepared from B-PEI 3X diminish the cell uptake outcome, but not in 5X.
Cell toxicity and polyplex biocompatibility
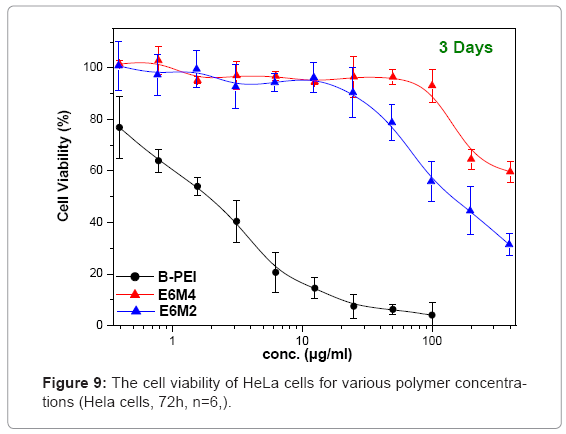
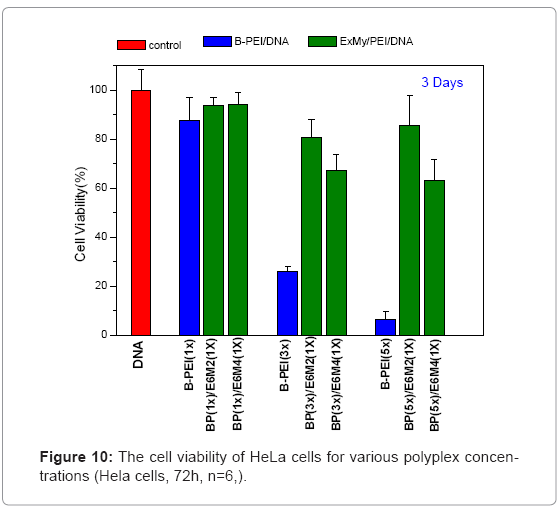
Figure 9 shows in vitro cytotoxicities of polymers, polycations BPEI, as a typical polymer for gene transfer, and PEOz-b-PMAA (E6M2 and E6M4) at various concentrations after 72 h incubation on HeLa cultures. This study treated the wells containing only media without polycations as positive controls and set them at a cell viability of 100%, calculating relative cell viability as [Abs]sample/[Abs]control x 100. The B-PEI exhibited stronger cytotoxicity with an IC50 = 2.5 μg/ml compared to PEOz-b-PMAA with IC50 = about 150 μg/ml for E6M2, and that of E6M4 was higher than 500 μg/ml. The cell viability of PEOzb- PMAA declined as the PMMA percentage increased. It may be due to E6M4 has more negative charge from PMMA segment than E6M2 which make E6M4 hard to be absorbed by cell. Figure 10 shows polyplex cytotoxicities with B-PEI and PEOz-b-PMAA at various concentrations after 72 h incubation on HeLa cultures. To obtain the difference in biocompatibility between B-PEI/DNA and the PEOz-b-PMAA ternary polyplex, this study selected the ternary polyplex with a zeta potential of about 0~10mV for the HeLa cell viability test and compared it with the B-PEI/DNA polyplex. The cell culture incubated with DNA was the control. The results indicated cell viability: B-PEI 1X > B-PEI 3X > B-PEI 5X. After we covered it with PEOz-b-PMAA, the cell viability of these ternary polyplexes maintained high viability independent of the content composition of the core structure. Decreased toxicity resulted from shielding the positively charged polymer in the core by the PEOz hydrophilic shell structure. The previously mentioned results indicated a slight positive charge of the PEOz does not damage the cells as in B-PEI.
Polyplex morphology by TEM observation
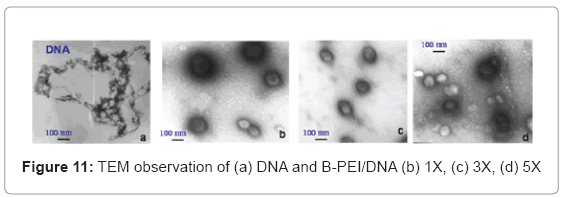
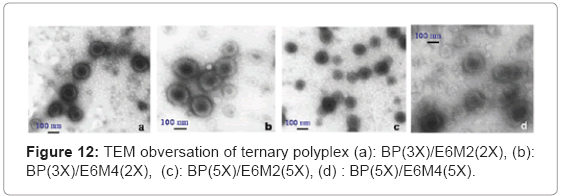
TEM observed the polyplex morphology. The study incubated the polyplexes in the phosphate buffer (PH=7) and succinate buffer (PH=5) solutions at room temperature for 1 h to observe the PH-sensitive behavior. The TEM images in Figure 11(a) show that free DNA has a random coil structure. Figure 11(b), (c) and (d) show the B-PEI/DNA polyplex does not have a core-shell, but a global structure. In this case, when the B-PEI wraps the DNA to form a compact particle, the DNA cannot react with uranyl acid (UA) easily. The images of B-PEI/DNA demonstrate a white contrast in the particle center. Unlike B-PEI, E6M2/DNA and E6M4/DNA polyplexes form an obvious core-shell micelle structure as shown in Figures 12(a)~(d). The inner core, P(EOz/EI) complex with DNA, is negatively stained by UA and exhibits a dark image, where the nuclei acid from the DNA bond with UO2 (Ac) + due to UA dissociation [16]. The outer shell, the PEOz hydrophilic segment, provides less contrast. The hydrophilic and neutral charge segment PEOz of the shell may protect polyplexes from immune system destruction.
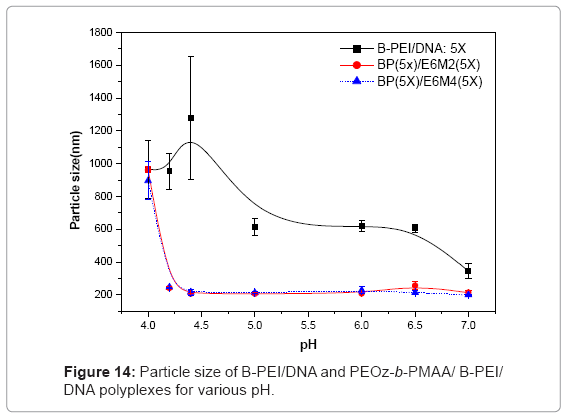
Figures 13(a) ~ (d) show the PH-sensitivity of the ternary polyplex. The PH response of the polyplex was carried out at PH 5. The random coil structure from DNA interconnected with the broken particles and the core-shell structures broke under acid conditions. The ruptured ternary polyplex formed a gel-like state and stuck together. The PH responsive PEOz-b-PMAA produced by hydrogen bonding in an acidic environment may produce this state. The particle size showed the same results (Figure 14). Different from the B-PEI/DNA polyplex, the particle size of the ternary polyplex showed a great rise in the acidic environment. In short, the PH-sensitivity from PEOz-b-PMAA of the ternary polyplex may help DNA release from the polyplexes in the endosome after cell uptake.
Cellular uptake of polyplexes
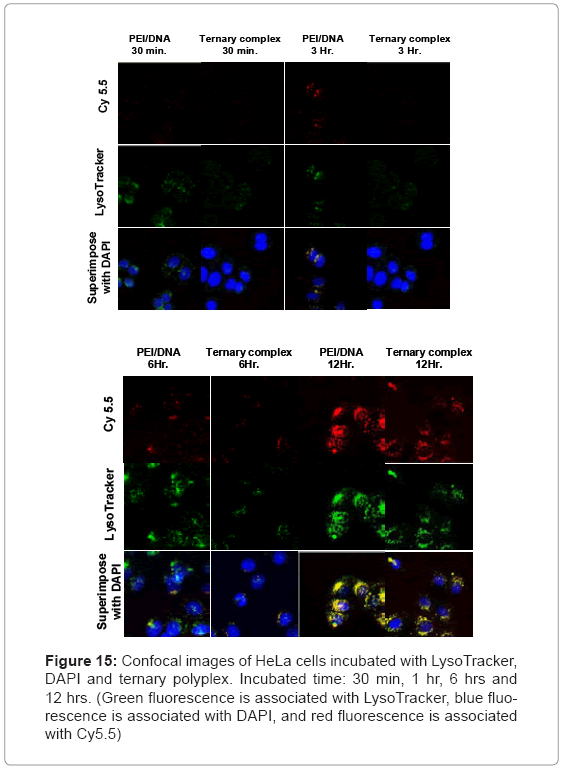
Figure 15 shows the time-dependent fluorescence intensity of Cy5.5, LysoTracker, and DAPI after incubating B-PEI/DNA polyplexes and the ternary polyplex with HeLa cells. The current study used the LysoTracker and DAPI molecules as indicators in the acidic compartment and nucleus [17]. The CLSM traced the cell uptake process. The fluorescence signals of DAPI revealed a blue color, the signals of Lysotracker revealed a green color, and Cy5.5 was red. For B-PEI/DNA, the Cy5.5 signals intensified around the HeLa cell periphery. The B-PEI remained in the cell nucleus after 6 h. The cell uptake progress for the ternary polyplex was slower. Observations showed significant amounts of Cy5.5 signals at 6 h, and the ternary polyplex entered the cell nucleus at 12 h. The B-PBI/DNA polyplex showed a more obvious signal after 6 h, compared with the ternary polyplex because of the higher positive charge density on B-PBI for enhancing cell uptake.
Conclusion
This study synthesized the PH-responsive diblock copolymer PEOzb- PMAA by polymerizing the MMA monomer with the macroinitiator (PEOz)2-ABCPA. The study successfully prepared the ternary polyplex based on PEOz-b-PMAA and B-PEI, and evaluated the physicochemical and biological characteristics. The results showed the ternary polyplex efficiently covered up the positive charge of the B-PEI/DNA precomplex and offered higher cell viability than the B-PEI/DNA polyplex. The ternary polyplex formed a clear core-shell structure by complexing with DNA under TEM observation. These polyplexes show PH-sensitivity to destroy core-shell structures and gene release in acidic buffers at a PH of 5. The non-viral gene carrier based on polyion PEOz-b-PMAA creates an optimal particle size and zeta potential for the polyplex, and helps DNA release from polyplexes with PH-sensitivity. Accordingly, the ternary polyplex may have much potential in non-viral gene therapy.
References
- Han SO, Mahato RI, Sung YK, Kim SW (2000) Development of biomaterials for gene therapy. Mol Ther 2: 302-317.
- Tang MX, Szoka FC (1997) The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Ther 4: 823-832.
- Luo D, Saltzman WM (2000) Synthetic DNA delivery systems. Nat. Biotechnol 18: 33-37.
- Pollard H, Remy JS, Loussouarn G, Demolombe S, Behr JP, et al. (1998) Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J Biol Chem 273: 7507-7511.
- Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, et al. (1996) A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: Polyethylenimine. Hum Gene Ther 7: 1947-1954.
- Boussif O, Lezoualc¢h F, Zanta MA, Mergny MD, Scherman D, et al. (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc Natl Acad Sci USA 92: 7297-7301.
- Brazeau GA, Attia S, Poxon S, Hughes JA (1998) In vitro myotoxicity of selected cationic macromolecules used in non-viral gene delivery. Pharm Res 15: 680-684.
- Takakura Y, Hashida M (1996) Macromolecular carrier systems for targeted drug delivery: pharmacokinetic considerations on biodistribution. Pharm Res 13: 820-831.
- Zalipsky S, Hansen CB, Oaks JM, Allen TM (1996) Evaluation of blood clearance rates and biodistribution of poly(2-oxazoline)-grafted liposomes. J Pharm Sci 85: 133-137.
- Ging-Ho Hsiue, Hou-Zen Chiang, Chau-Hui Wang, Tzuoh-Min Juang, (2006) Nonviral Gene Carriers Based on Diblock Copolymers of Poly(2-ethyl-2-oxazoline) and Linear Polyethylenimine. Bioconjugate Chem 17: 781-786.
- G Decher J, D Hong (1991) Makromol. Chem Macromol Symp 46: 321.
- G Decher, Fuzzy (1997) Nanoassemblies: Toward Layered Polymeric Multicomposites. Science 277: 1232.
- Vladimir S Trubetskoy, Aaron Loomis, James E Hagstrom, Vladimir G Budker1, Jon A Wolff (1999) Nucleic Acids Research 27: 3090-3095.
- Yoshiyuki K (2003) Novel poly(ethylene glycol) derivatives having carboxylic acid pendant groups: Syntheses and their enhancing effect on non-viral gene transfection systems. Sci Polymer Edn 14: 515-531.
- JL Thomas, DA Tirrell (2000) Polymer-Induced Leakage of Cations from Dioleoyl Phosphatidylcholine and Phosphatidylglycerol Liposomes. J Control Rel 67: 203.
- Huxley HE, Zubay G (1961) Preferential Staining of Nucleic Acid-Containing Structures for Electron Microscopy. Biophys Biochem Cytol 11: 273-296.
- Burdette SC, Walkup GK, Spingler B, Tsien RY, Lippard SJ (2001) Fluorescent Sensors for Zn2+ Based on a Fluorescein Platform: Synthesis, Properties, and Intracellular Distribution. J Am Chem Soc 123: 7831-7841.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 15761
- [From(publication date):
July-2011 - Nov 01, 2025] - Breakdown by view type
- HTML page views : 11052
- PDF downloads : 4709