Research Article Open Access
Surface Plasmon Resonance Immunosensor for Detection of PB1-F2 Influenza A Virus Protein in Infected Biological Samples
Jasmina Vidic*, Christophe Chevalier, Ronan Le Goffic, Anna Miodek, Christiane Bourdieu, Charles-Adrien Richard, Mohammed Moudjou and Bernard Delmas
Virologie et Immunologie Moléculaires, Institut National de la Recherche Agronomique, France
- *Corresponding Author:
- Jasmina Vidic
Virologie et Immunologie Moléculaires
Institut National de la Recherche Agronomique
78350 Jouy en Josas, France
Tel: +0033134652623
Fax: +0033134652621
E-mail: jasmina.vidic@jouy.inra.fr
Received date: February 22, 2013; Accepted date: April 02, 2013; Published date: April 04, 2013
Citation: Vidic J, Chevalier C, Le Goffic R, Bourdieu C, Richard CA, et al. (2013) Surface Plasmon Resonance Immunosensor for Detection of PB1-F2 Influenza A Virus Protein in Infected Biological Samples. J Anal Bioanal Tech S7:006. doi: 10.4172/2155-9872.S7-006
Copyright: © 2013 Vidic J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
The detection and evaluation of concentration of influenza virus proteins in biological samples is critical in a broad range of medical and biological investigations regarding the concern over potential outbreaks of virulent influenza strains in animals and humans. This paper describes a sensitive, label-free approach for the detection of a virulence factor PB1-F2. PB1-F2 is a small, 90 amino acid long polypeptide expressed in influenza A viruses, which generally exacerbate virus pathogenicity. The developed immunosensoris based on a non-the-chipcovalently immobilized specific monoclonal anti-PB1-F2 antibody and a SPR technology. The immunosensor was calibrated using purified full length PB1-F2 protein. Itdetected PB1-F2 with the linear range extended from 10 to 500 nM, repeatability of 5% for 500 nM PB1-F2 and showed saturationof protein concentrations higher than 1 μM. The sensor can quantify PB1-F2 in its monomeric form but not when its oligomerization was induced by preincubation in 0.05% SDS. The immunosensor was successfully applied in the detection and quantification of PB1-F2 in infected mouse lungs and cell lines, providing temporal expression profiles of PB1-F2 during viral infection. In lungs of infected mice, the influenza virus structural nucleoprotein NP was detected in parallel using a specific anti-NP antibody. This parallel detection of PB1-F2 and NP suggests that applied sensor chip technology may be amenable to an arrow immunosensor for simultaneous detection of all known influenza virus proteins in infected tissues and cells.
Keywords
Biosensing; Influenza A virus; PB1-F2; A549 cells; U937 cells; Mouse lung
Abbreviations
AU: Absorbance Unit; IAV: Influenza A Virus; NP: Nucleoprotein; PrP: Prion Protein; RU: arbitrary Resonance Unit; SDS: Sodium Dodecyl Sulfate; SPR: Surface Plasmon Resonance; ThT: ThioflavinT; WT: Wild Type
Introduction
Every year, seasonal Influenza A virus (IAV) affects millions of people leading to illness and death. This virus causes a severe respiratory tract infection in high-risk populations such as elderly people, young children and patients with chronic lung and heart diseases. IAV was responsible for three main pandemics of the last century in 1918, 1957 and 1968. The World Health Organization is concerned about the highly virulent strains of IAV, such as H5N1 avian viruses which has about 60% lethality rate [1]. Increasing numbers of confirmed human cases for avian influenza A (H5N1) were reported to WHO between 2003 and 2011. In 2009, the novel swine IAV H1N1 caused the first pandemy of the XXI century, with an estimated number of about 12,500 H1N1-related deaths.
IAV are enveloped viruses belonging to the Orthomyxoviridae family. Their genomes are composed of eight segments of negative strand RNA encoding 13 known viral proteins [2-4]. The highly abundant structural IAV proteins are detectable in cell lysates or in purified viruses, by polyacrylamide gel electrophoresis. However, less abundant and non-structural proteins were discovered by screen dipitolously or deep analysis of the genome open reading frames and then further characterized using specific antibodies or mass spectrometry [5]. In 2001 Cheng et al. [3] discovered a new influenza virus protein, encoded by the PB1 segment from an alternative +1 open reading frame, named PB1-F2 for PB1-frame 2. The PB1-F2 was found to be a small protein of about 90 amino acids. The specific contribution of PB1-F2 to virulence is still uncertain despite numerous studies associating PB1-F2 with apoptosis induction [6-8], and with enhanced risk of secondary bacterial infections [9]. Using a model of mouse infection Zamarin et al. [8] demonstrated that wild type IAV displays a higher pathogenicity than its PB1-F2 knock-out counterpart.
PB1-F2 is considered as one of the factors that contributed to the virulence of the most lethal IAV strain, Spanish influenza, which killed between 20 and 40 million of people in 1918. A single N66S mutation present in PB1-F2 of 1918 pandemic strain was sufficient to transform a strain of moderate virulence into a highly pathogenic virus, as shown in mice [10]. Since 1947, human H1N1 strains have been encoded truncated PB1-F2. These viruses lack the PB1-F2-mediated capacity of apoptosis induction, and interestingly they usually cause mild symptoms in comparison with counterparty H3N2 seasonal strains that express a full length PB1-F2 protein. This suggests a relationship between virulence of IAV, ability to induce apoptosis and PB1-F2 expression.
PB1-F2 has a short half-life and it seems to be rapidly degraded within a few hours [3,11]. PB1-F2 was shown to target on both inner and outer protein mitochondrial membranes, influencing virally induced apoptosis [3,8]. PB1-F2-induced apoptosis is mediated by mitochondrial permeabilization followed by a cytochrome C release. In addition, PB1-F2 of several IAV strains was found to co-localize with PB1 in the nucleus of infected cells and to enhance viral polymerase activity [12]. However, complete elucidation of molecular mechanisms of PB1-F2 participation in viral infection remains unknown and demands new study approaches.
In vitro studies showed that PB1-F2 shares some properties with small intrinsically disordered proteins [13]. We recently produced full length PB1-F2 variants from seven IAV strains, including the one of the highly pathogenic 1918 virus [13]. None of the PB1-F2 variants acquires a secondary structure in aqueous solutions, but all adopt β-sheet conformation in a hydrophobic environment. Furthermore, PB1-F2 can oligomerize to form amyloid fibers in the presence of liposomes or SDS at concentrations below its critical micelle concentration. In addition, PB1-F2 was shown to form amyloid fibers in IAV infected cells [11,13].
To reveal the expression of PB1-F2 in IAV infections, a sensitive method for its detection and quantification in biological samples is needed. Here we used surface plasmon resonance (SPR) technology to develop an immunosensor for the detection of PB1-F2. The sensor is based on the specific anti-PB1-F2 monoclonal antibody that recognizes PB1-F2. In optimized conditions the immunosensor was highly sensitive and reproducible, and allowed the estimation of PB1-F2 concentration at a nanomolar range in complex biological medium. We believe that the developed sensor has potential as a valuable diagnostic and analytical tool for influenza virus studies.
Experimental
Antibodies
Rabbit hybridoma monoclonal anti-PB1-F2 antibodies were kind gifts from Dr J.F. Vautherot (INRA). The antibodies were produced against the full-length PB1-F2 His-tag of WSN virus strain. Antinucleoprotein (anti-NP) antibody was purchased from Santa Cruz (Invitrogen, France) Influenza A NP (968).
Generation of recombinant influenza virus
We used influenza A/WSN/1933 (H1N1) in this study. Viral stocks of wild-type andPB1-F2 knockout virus mutant were produced as previously described [13,14].
Protein expression and purification
Full-length PB1-F2His-tag of WSN influenza virus was expressed and purified as described previously [13]. Briefly, the gene encoding PB1-F2 was cloned into the E.coli expression vector pET 22b+ and expressed by IPTG induction in the BL-21 Rosetta DE3 E.coli strain. After cell lysis the protein was purified from inclusion bodies on a Ni-Sepharose column by heterogeneous phase renaturation, as explained previously [13,14]. PB1-F2 HisTag protein was lyophilized and stored at -20°C. Prior to analysis lyophilized protein powder was dissolved in 10 mM sodium-acetate buffer, pH 5. Final protein concentration was determined by measuring optical density at 280 nm and concentration was established using extinction coefficient deduced from its composition of 28990 M-1 cm-1. The monomeric form of PB1-F2 was stable for several weeks in 10 mM sodium acetate buffer, pH 5 at 4°C. Expression and purification of nucleoprotein (NP) was described previously [15]. Briefly, the full-length NP of H1N1 (strain A/WSN/33) with a His-tag on its N-terminal was expressed E.coli BL21 (DE3) and purified by a two-step procedure, firstly by IMAC-Ni2+ affinity chromatography, and then by size-exclusion chromatography.
We used Shadoo [16] and the N-terminal of Prion (ARQ 23-133) [17], proteins as controls.
Cell culture
The human alveolar epithelial cell line A549 and the human promonocytic cell line U-937 were purchased from the American Type Culture Collection (Manassas, VA). Cells were propagated and maintained in RPMI 1640 medium (Lonza) supplemented with 10% fetal calf serum, 2 mM L-glutamine, 100 IU/ml penicillin and 100 μg/ ml streptomycin, according to the American Type Culture Collection recommendations. Cells were maintained at 37°C in a 5% CO2 incubator. For infections, cells were washed with fetal calf serum-free medium and incubated with WSN virus at 0.1 multiplicity of infection for 1 h at 37°C. Infected cells were then incubated at 37°C with complete fetal calf serum until collection.
Ethics statement
This study was carried out in accordance with INRA guidelines in compliance with European animal welfare regulation. The protocols were approved by the Animal Care and Use Committee at CRJ under (COMETHEA; protocol number 12-060) relevant institutional (DSV, permit number: A78-167) and INRA “Santé Animale” department guidelines. All experimental procedures were performed in a Biosafety level 2 facility.
Mice and viral infection
Female C57Bl/6 mice were purchased from the Centre d’Elevage R. Janvier (Le Genest Saint-Isle, France). Mice were housed in the animal facility of the Jouy-en-Josas campus under standard conditions with at a constant temperature of 24°C with 12:12 daylight cycle in pathogenfree conditions. Food and water were provided ad libitum. For infection experiments, mice of around 8 weeks of age were housed in cages inside stainless steel isolation cabinets that were ventilated under negative pressure with HEPA-filtered air. Mice were anesthetized by a mixture of ketamine and xylazine (1 and 0.2 mg per mouse, respectively) and infected intranasally with 50 μl of PBS containing 1×106, 5×105, 2.5×105, 1.5×105 or 5×104 plaque forming units (PFU) of IAV. Mice were killed at different time points, broncho-alveolar lavage fluids and lungs were then collected as previously described [14]. Quantification of sensor response was done on lung homogenates from 3-4 different mice in each group.
Sensor design
Surface Plasmon resonance (SPR) analysis was performed at 25°C using the Biacore 3000 (GE Healthcare). To immobilize an anti-PB1-F2 antibody onto the surface of a CM5 commercial chip (Biacore) via amino-link, the carboxymethyldextran surface of the CM5 was activated using an Amino coupling kit (Biacore). Anti-PB1-F2 antibody diluted in 10 mM sodium acetate buffer, pH 5, was injected onto the activated surface giving up to 3000 resonance units (RU). After immobilization of the antibody, the remaining activated surface was blocked by an Ethanolamine (1 M). To find a calibration curve of the sensor, purified recombinant PB1-F2 was injected at flow rate of 25 μl/min for 2 min. SPR signal from the uncoupled surface (activated, and inactivated by Ethanolamine) was used as a reference for each protein concentration. Regeneration of the anti-PB1-F2 antibody was achieved by a 2 min injection of 10 mM Glycine buffer, pH 2.2. All measurements were performed at least in triplicate. Obtained sensorgrames were analyzed by BIA evaluation software (Biacore, GE Healthcare).
ThT fluorescent measurements
Thioflavin T (ThT) measurements were performed on a Jacso FR- 6200 spectrofluorimeter with 1 mm×10 mm optical path-length quartz cuvettes at 20°C. Fibrilization of PB1-F2 was obtained by mixing 10 μM PB1-F2 with 0.05% SDS in 10 mM sodium-acetate buffer, pH 5. Solutions were preincubated at room temperature for 1 hour before measurements. Then, fibers formed were marked with ThT (10 μM). Excitation was performed at 432 nm, and measurement of fluorescent emission was performed from 450 nm to 620 nm.
Electron microscopy
A 10 μL PB1-F2 fibril preparation was collected on Formvar/carboncoated 200-mech copper grids (Agar Scientific). After deposition of the suspension, grids were washed twice for 1 min with phosphate buffer saline, pH 7.4, and negatively stained by floating on 10 μl drops of 2% (w/v) uranyl acetate (Sigma) for 1 min. The grids were air-dried before observation under Philips EM12 electron microscope at an 80-kV exciting voltage.
Results and Discussion
PB1-F2 detection and analytical parameters of the immunobiosensor
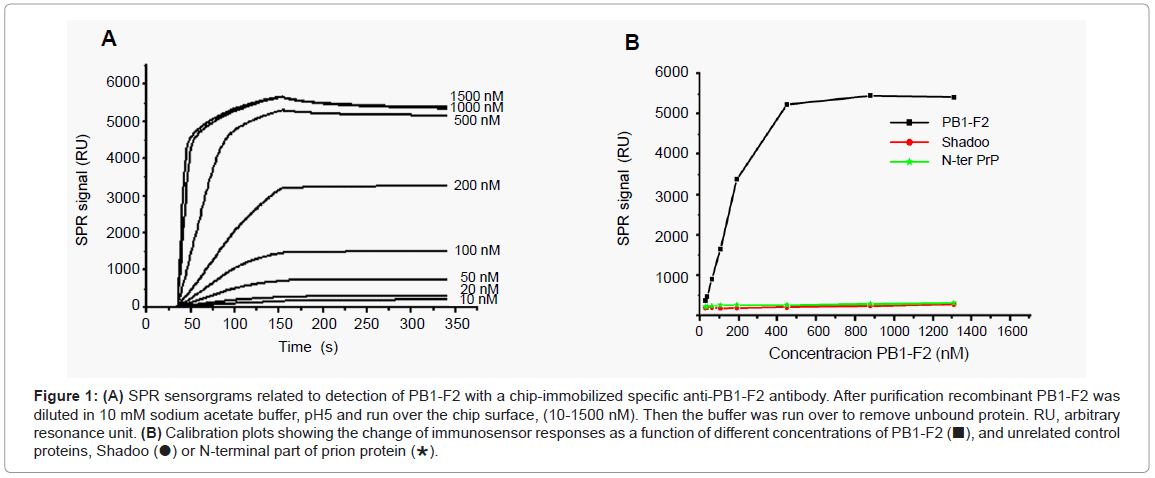
A SPR biosensor assay based on the specific monoclonal anti- PB1-F2 antibody was developed to allow the quantification of PB1-F2. The antibody was attached covalently to the surface of the CM5 chip by amino link. The sensitivity and selectivity of the immunobiosensor was explored with a full length recombinant PB1-F2 that was injected at various concentrations over immobilized anti-PB1-F2 antibody. To avoid PB1-F2 aggregation the interaction of the protein with the chip surface was followed in sodium acetate buffer, pH 5 since in this buffer PB1-F2 is monomeric [13]. Sensorgrams presented in Figure 1A show that upon PB1-F2 injection the antigen-antibody complex was formed giving rise to an increase in SPR signal. A calibration curve corresponding to the variation of SPR signal versus concentration of injected PB1-F2 is shown in Figure 1B. The calibration curve indicates that the immunosensor exhibits a linear relationship with PB1-F2 concentration ranging from 10 nM to 500 nM with a threshold of 10nM, and shows signal saturation for PB1-F2 concentrations higher than 1 μM.
Figure 1: (A) SPR sensorgrams related to detection of PB1-F2 with a chip-immobilized specific anti-PB1-F2 antibody. After purification recombinant PB1-F2 was diluted in 10 mM sodium acetate buffer, pH5 and run over the chip surface, (10-1500 nM). Then the buffer was run over to remove unbound protein. RU, arbitrary resonance unit. (B) Calibration plots showing the change of immunosensor responses as a function of different concentrations of PB1-F2 (■), and unrelated control proteins, Shadoo (●) or N-terminal part of prion protein (∗).
The selectivity of the biosensor towards PB1-F2 was tested with nonrelated proteins such as recombinant Shadoo and the N-terminal part of prion protein. Shadoo and N-terminal prion protein were chosen for their molecular weight and total charge similar to those of PB1-F2: PB1-F2 is a protein of 11 kDa, Shadoo of 12 kDa, and N-terminal prion protein of 10.8 kDa; all three proteins have theoretical pI of about 10. In addition, Shadoo protein was His-tagged, as was the PB1-F2. Figure 1B shows that no significant SPR signal increase was observed with either Shadoo or N-terminal prion proteins in a concentration range from 10 nM to 1.5 μM, demonstrating the specificity of PB1-F2 detection.
The reproducibility of SPR measurement for PB1-F2 detection was tested on 6 biosensors fabricated with the same procedure on new flow cells of CM5 chips, a RSD value of 5% was obtained for 500 nM PB1-F2.
No detection of PB1-F2 fibers
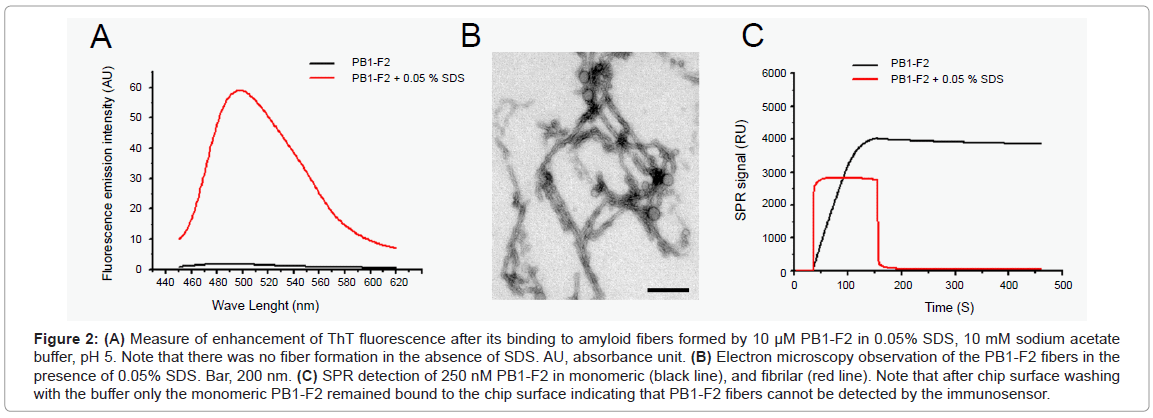
We have previously shown that PB1-F2 can polymerize to form amyloid fibers and different types of oligomers in infected cells in the vicinity of cellular membranes [13]. We, thus, checked for the detection of oligomerized forms of PB1-F2 by the immunosensor. The PB1-F2 fibrilization was obtained by incubating PB1-F2 in 0.05% SDS /sodium acetate buffer, pH 5. SDS is an ionic detergent usually used to mimic biological membranes [18]. The oligomerization of PB1-F2 to amyloid fiber was verified by ThT staining. ThT specifically binds β-sheet rich structures like amyloid fibers and displays an enhanced fluorescence and a characteristic red shift of its emission spectrum. Figure 2A shows that ThT detected PB1-F2 amyloid fibers in protein preparation incubated with SDS. In contrast, no ThT fluorescence was detected in PB1-F2 buffer solution without SDS. The amyloid fiber formation was confirmed by electron microscopy observation of negatively stained preparations since fibers of several hundred nm length were detected in 0.05% SDS protein solution (Figure 2B).
Figure 2: (A) Measure of enhancement of ThT fluorescence after its binding to amyloid fibers formed by 10 μM PB1-F2 in 0.05% SDS, 10 mM sodium acetate buffer, pH 5. Note that there was no fiber formation in the absence of SDS. AU, absorbance unit. (B) Electron microscopy observation of the PB1-F2 fibers in the presence of 0.05% SDS. Bar, 200 nm. (C) SPR detection of 250 nM PB1-F2 in monomeric (black line), and fibrilar (red line). Note that after chip surface washing with the buffer only the monomeric PB1-F2 remained bound to the chip surface indicating that PB1-F2 fibers cannot be detected by the immunosensor.
Next, a 250 nM PB1-F2 solution, either its monomeric or fibrillated form, were run over two identical immunosensor flow cells carrying anti-PB1-F2 antibody. As expected, the immunosensor detected the monomeric PB1-F2 giving about 3700 RU signal increase (Figure 2C). However, PB1-F2 fiber injection gave no signal increase as shown in Figure 2C. This finding strongly suggests that anti-PB1-F2 antibody used here cannot detect PB1-F2 in its amyloid fiber form, probably since the specific epitope is hidden in the fiber structure. In consequence, the immunobiosensor cannot quantify the total amount of protein, but only the free monomeric PB1-F2.
PB1-F2 expression profiles in infected cells
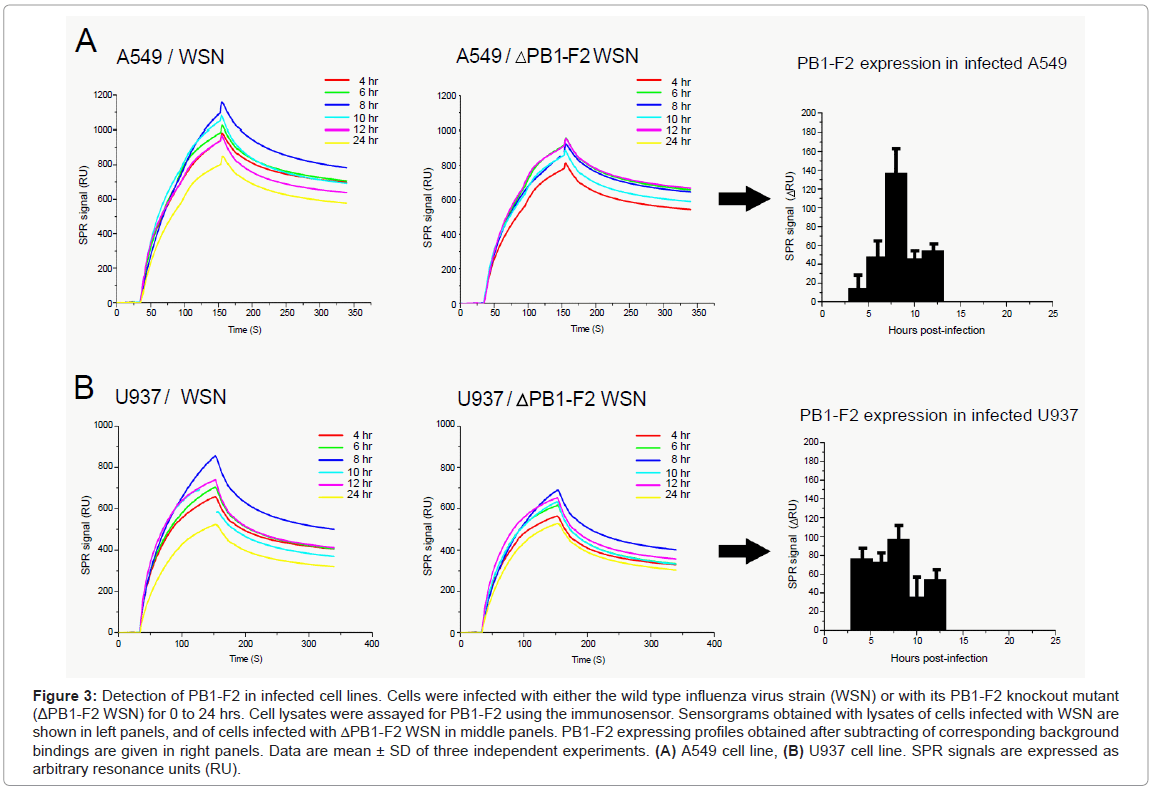
Human alveolar epithelial A549 and monocyte U937 cell lines were infected with a recombinant wild type influenza virus (WSN), or with a mutant that was knock out for the PB1-F2 gene expression (ΔWSN). Cells were collected and lysed at various time points post infection. Cellular lysates obtained by successive freeze-thawed were adjusted to a final protein concentration of 0.1 mg/ml and then run over anti- PB1-F2 antibody immobilized on flow cells (Figure 3). Lysates of ΔWSN-infected cells were used for signal background estimation. Indeed, previous reports indicate that there is no significant difference of progeny virus titers between wild type and PB1-F2 knock out virus upon infection of various cell lines or mouse lung [3,8,13,14]. The second negative control was lysates of non-infected cells (data not shown). The relevant SPR response signals of PB1-F2 binding to the biosensor surface were obtained by subtracting the corresponding SPR responses from lysates of cells infected with mutant virus from those infected with the wild type virus (Figures 3A and 3B). This elimination of non-specific binding allowed the estimation of PB1-F2 concentration at various time periods after the virus infection.
Figure 3: Detection of PB1-F2 in infected cell lines. Cells were infected with either the wild type influenza virus strain (WSN) or with its PB1-F2 knockout mutant (ΔPB1-F2 WSN) for 0 to 24 hrs. Cell lysates were assayed for PB1-F2 using the immunosensor. Sensorgrams obtained with lysates of cells infected with WSN are shown in left panels, and of cells infected with ΔPB1-F2 WSN in middle panels. PB1-F2 expressing profiles obtained after subtracting of corresponding background bindings are given in right panels. Data are mean ± SD of three independent experiments. (A) A549 cell line, (B) U937 cell line. SPR signals are expressed as arbitrary resonance units (RU).
Interestingly, the expression profiles of PB1-F2 obtained in A549 and U937 cells were not identical. In U937 cells, the maximal accumulation of PB1-F2 was reached within 4 hrs post infection staying almost constant in the following 8 hrs. In contrast, PB1-F2 expression in A549 cells starts within 6 hrs post infection and then accumulates for up to 10 hrs. Regarding the calibration curve from Figure 1 the PB1-F2 concentration in A549 cells can be estimated to reach 100-200 nM, while in U937 cells PB1-F2 reached a maximum of 50-100 nM. In both cell lines, no monomeric PB1-F2 was detected at 24 hours post infection. The absence of PB1-F2 24 hrs post infection could originate from transient protein expression and its short half-life. The transient expression of PB1-F2 was previously shown by Western blot analysis on infected cells using antisera or a polyclonal antibody specific to PB1-F2 [3,8,11,12] while protein half-life was estimated to be between 30 minutes [3] and 1 hour [8]. However, the possibility that PB1-F2 formed amyloid fibers when accumulated in infected cells cannot be ruled out. Indeed, we previously detected PB1-F2 amyloid fibers in both U937 and A549 12 hours post infection [11,14]. This time period overlaps with a fall in concentration of monomeric PB1-F2. To distinguish between these two possibilities biosensing of PB1-F2 amyloid fibers in parallel to monomer quantification is needed.
Mazur et al. [12] have shown that in A549 cells PB1-F2 can localize to the mitochondria, cytoplasm and nucleus of infected cells. The multiple cellular localization of PB1-F2 suggests the protein has a multifunctional role. They have shown that besides promoting apoptosis, PB1-F2 also increases polymerase activity of viral PB1 in epithelial cells [12]. In contrast, early studies on the functional apoptosis-promoting role of PB1-F2 in immunological cells, such as monocytes, suggested only mitochondrial localization of PB1-F2 [3]. Moreover, Zamarin et al. [8] proposed that in infected animals PB1-F2 might selectively target immune cells to promote their apoptosis. Such apparently cell-dependent functional roles of PB1-F2 corroborate our finding that expression profiles of PB1-F2 are not identical in epithelial and immune cell lines. In consequence, it will be interesting to expand detection of PB1-F2 in other cell types to get insight into the specific expression profiles of PB1-F2 that may be further correlated with functional roles of PB1-F2.
PB1-F2 protein expression in lungs of infected mice
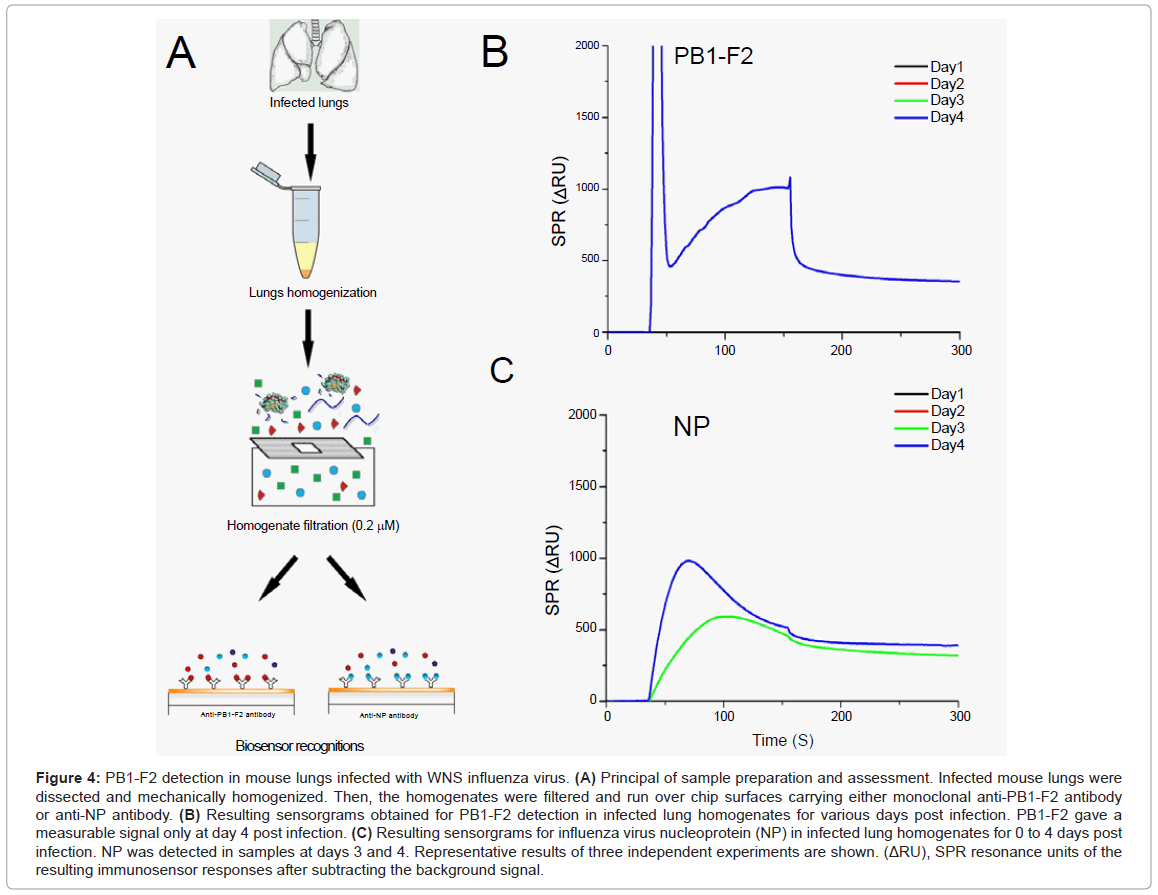
Finally, we attempt to detect PB1-F2 in the lungs of infected mice. For this, groups of 5 mice were infected with the mouse-adapted WSN strain. Mice were sacrificed at different time points and their lungs collected (Figure 4A). Before injection over the chip surface, lungs were mechanically homogenized and filtered to remove cell aggregates. This procedure was necessary to prevent damage to the microfluidic system device. It is important to note that this procedure would remove PB1-F2 amyloid fibres if they had formed in infected lungs.
Figure 4: PB1-F2 detection in mouse lungs infected with WNS influenza virus. (A) Principal of sample preparation and assessment. Infected mouse lungs were dissected and mechanically homogenized. Then, the homogenates were filtered and run over chip surfaces carrying either monoclonal anti-PB1-F2 antibody or anti-NP antibody. (B) Resulting sensorgrams obtained for PB1-F2 detection in infected lung homogenates for various days post infection. PB1-F2 gave a measurable signal only at day 4 post infection. (C) Resulting sensorgrams for influenza virus nucleoprotein (NP) in infected lung homogenates for 0 to 4 days post infection. NP was detected in samples at days 3 and 4. Representative results of three independent experiments are shown. (ΔRU), SPR resonance units of the resulting immunosensor responses after subtracting the background signal.
In order to compare the expression profile of PB1-F2 to a viral structural protein NP, lung homogenates were injected over two flow cells, one carrying anti-PB1-F2 antibody and other anti-NP antibody. PB1-F2 protein was detected at day 4, while NP was detected at days 3 and 4 (Figures 4B and 4C). We examined the expression profile of PB1-F2 for days 1-4 post infection as the WSN strain induced 80% lethality in infected mice at day 5 post infection [14]. sensorgram grams given in Figure 4 result from subtracting sensorgram grams of corresponding negative controls (obtained by injection of lung homogenates from mice infected with Δ PB1-F2WNS, or lungs of non-infected mice) from those obtained from mice infected with wild-type virus. Signal intensities allowed estimation of protein concentrations to range around 26 pmol per mg of lung tissue for PB1-F2 and from 500 to 700 pmol per mg of lung tissue for NP (calibration curve obtained with purified monomeric NP is not shown). Again we cannot rule out the possibility that a fibrilar form of PB1-F2 is involved in viral pathogenesis, but it is evident that the highest PB1-F2 monomeric concentration is reached when immunopathological disorders take place.
The possibility to detect two influenza virus proteins in parallel in infected tissue using the SPR technology opens new opportunities for immediate applications, such as a sensitive protein array for in parallel detection of all virus proteins in infected tissues, and a development of new diagnostic tools for influenza virus infections.
Conclusions
Biological biosensors allow rapid and specific detection of biological molecules and could replace many of the analytical methods currently used in biochemistry and cellular biology. Here we report that influenza virus protein PB1-F2 can be directly detected in biological samples using a specific monoclonal anti-PB1-F2 antibody in combination with SPR-based sensor. We have shown that immunosensors can evaluate the PB1-F2 concentration in infected cells and tissues. The limitation of this device is that it cannot detect the polymerized form of PB1-F2 probably because the specific epitopes are not accessible in this structure. The SPR assay is simple, does not need any sample labeling, and the PB1-F2 concentration is available within 10 minutes. In addition, this specific SPR assay has notably higher sensitivity compared with the Western-blot or immunospectrometry. Thus, we believe that this new immunobiosensor has great potential for use in both biological and medical investigations of influenza virus pathologies.
Summary
The detection and evaluation of concentration of influenza virus proteins in biological samples is critical in a broad range of medical and biological investigations regarding the concern over potential outbreaks of virulent influenza strains in animals and humans. This paper describes a sensitive, label-free approach for the detection of a virulence factor PB1-F2. PB1-F2 is a small unstructured protein, expressed in influenza A viruses. The role of PB1-F2 in viral infection is not well understood but, many studies have showed that it exacerbates virus pathogenicity. The developed immunosensor is based on an on-thechip covalently immobilized specific monoclonal anti-PB1-F2 antibody and a surface plasmone resonance technology. The immunosensor was calibrated using purified full length PB1-F2 protein. Linear range of sensor detection, sensor repeatability and PB1-F2 concentration giving signal saturation were determined. The sensor can quantify PB1-F2 in its monomeric but not in its oligomerizating form. The immunosensor was successfully applied in the detection and quantification of PB1-F2 in infected mouse lungs and cell lines, providing time expression profiles of PB1-F2 during viral infection. We believe that this new immunobiosensor has great potential for use in both biological and medical investigations of influenza virus pathologies.
Acknowledgement
We thank Dr Rachel Young (Institut Gustave Roussy, Villejuif, France) for critical reading of the manuscript and Dr Jean-François Vautherot (INRA, Nouzilly, France) for providing monoclonal anti-PB1-F2 antibody.
References
- World Health Organization (2011) Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003-2011.
- Palese P (1977) The genes of influenza virus. Cell 10: 1-10.
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, et al. (2001) A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7: 1306-1312.
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, et al. (2012) An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337: 199-204.
- Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P (2008) Cellular proteins in influenza virus particles. PLoS Pathog 4: e1000085.
- Chanturiya AN, Basañez G, Schubert U, Henklein P, Yewdell JW, et al. (2004) PB1-F2, an influenza A virus-encoded proapoptotic mitochondrial protein, creates variably sized pores in planar lipid membranes. J Virol 78: 6304-6312.
- Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW (2003) The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol 77: 7214-7224.
- Zamarin D, García-Sastre A, Xiao X, Wang R, Palese P (2005) Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog 1: e4.
- McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, et al. (2007) Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2: 240-249.
- Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P (2007) A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 3: 1414-1421.
- Le Goffic R, Bouguyon E, Chevalier C, Vidic J, Da Costa B, et al. (2010) Influenza A virus protein PB1-F2 exacerbates IFN-beta expression of human respiratory epithelial cells. J Immunol 185: 4812-4823.
- Mazur I, Anhlan D, Mitzner D, Wixler L, Schubert U, et al. (2008) The proapoptotic influenza A virus protein PB1-F2 regulates viral polymerase activity by interaction with the PB1 protein. Cell Microbiol 10: 1140-1152.
- Chevalier C, Al Bazzal A, Vidic J, Février V, Bourdieu C, et al. (2010) PB1-F2 influenza A virus protein adopts a beta-sheet conformation and forms amyloid fibers in membrane environments. J Biol Chem 285: 13233-13243.
- Le Goffic R, Leymarie O, Chevalier C, Rebours E, Da Costa B, et al. (2011) Transcriptomic analysis of host immune and cell death responses associated with the influenza A virus PB1-F2 protein. PLoS Pathog 7: e1002202.
- Tarus B, Bakowiez O, Chenavas S, Duchemin L, Estrozi LF, et al. (2012) Oligomerization paths of the nucleoprotein of influenza A virus. Biochimie 94: 776-785.
- Watts JC, Drisaldi B, Ng V, Yang J, Strome B, et al. (2007) The CNS glycoprotein Shadoo has PrP(C)-like protective properties and displays reduced levels in prion infections. EMBO J 26: 4038-4050.
- Steunou S, Chich JF, Rezaei H, Vidic J (2010) Biosensing of lipid-prion interactions: insights on charge effect, Cu(II)-ions binding and prion oligomerization. Biosens Bioelectron 26: 1399-1406.
- Schwyzer R (1995) In search of the 'bio-active conformation'--is it induced by the target cell membrane? J Mol Recognit 8: 3-8.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 16504
- [From(publication date):
specialissue-2013 - Oct 19, 2025] - Breakdown by view type
- HTML page views : 11749
- PDF downloads : 4755