Special Issue Article Open Access
Ameliorating Risk: Culturable and Metagenomic Monitoring of the 14 Year Decline of a Genetically Engineered Microorganism at a Bioremediation Field Site
Alice C. Layton1*, Abby E. Smartt1, Oya Tekeli2, Archana Chauhan1, Steven Ripp1, Daniel E. Williams1, Whitney Burton1, Scott Moser1, Jana Phillips2, Anthony V. Palumbo2 and Gary S. Sayler1,21The University of Tennessee Center for Environmental Biotechnology, 676 Dabney Hall, Knoxville, Tennessee 37996, USA
2Oak Ridge National Laboratory, Biosciences Division, Oak Ridge, Tennessee, 37831, USA
- *Corresponding Author:
- Dr. Alice C. Layton
The University of Tennessee
Center for Environmental Biotechnology
676 Dabney Hall, Knoxville, Tennessee 37996, USA
Tel: 865-974-8072
Fax: 865-974-8086
E-mail: alayton@utk.edu
Received February 28, 2012; Accepted April 20, 2012; Published April 22, 2012
Citation: Layton AC, Smartt AE, Chauhan A, Ripp S, Williams DE, et al. (2012) Ameliorating Risk: Culturable and Metagenomic Monitoring of the 14 Year Decline of a Genetically Engineered Microorganism at a Bioremediation Field Site. J Bioremed Biodegrad S1:009 doi: 10.4172/2155-6199.S1-009
Copyright: © 2012 Layton AC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
In 1996, the first EPA sanctioned release of a recombinant microbe (Pseudomonas fluorescens HK44) into the subsurface soil environment was initiated in a replicated semi-contained array of soil lysimeters. With an aim to access the survivability/environmental fate of HK44, soil sampling was performed 14 years post release. Although after extensive sampling culturable HK44 cells were not found, qPCR and metagenomic analyses indicated that genetic signatures of HK44 cells still persisted in the soils, with genes diagnostic for the bioluminescent transposon carried by strain HK44 (luxA and tetA) being found at low concentrations (< 5000 copies/g). Additionally, metagenome analysis of lysimeter 2 using amplicon pyrosequencing showed that Burkholderia was more abundant in the sample extracted before storage at 4°C than after storage at 4°C (79% and 5.6% Burkholderia sequences, respectively).
Introduction
The first United States Environmental Protection Agency (EPA) approved application of a genetically engineered microorganism for bioremediation purposes occurred in 1996 at an intermediate scale (4 m deep × 2.5 m diameter), semi-contained, and replicated soil lysimeter site at the Oak Ridge National Laboratory, Oak Ridge, Tennessee [1]. This organism, Pseudomonas fluorescens HK44, was capable of degrading polycyclic aromatic hydrocarbons (PAHs) and contained the genes for bioluminescence (luxCDABE) downstream of the salicylate inducible nahR promoter on a plasmid (pUTK21) construct [2]. This plasmid allowed strain HK44 to degrade select twoto- three ring PAHs and other substituted PAHs while simultaneously emitting bioluminescent light, thereby permitting its use as a living bioreporter for the near real-time monitoring of bioremediation efficacy [3]. Strain HK44 was sprayed on to layered 10 cm soil lifts 1 m deep artificially contaminated with a PAH mixture of naphthalene, anthracene, and phenanthrene [4]. Bioluminescence from these soil-borne bioreporters was then monitored using fiber optic and photomultiplier tube (PMT) interfaces over the ensuing two years to establish the potential for bioreporters to serve as bioremediation process monitoring and control tools [1]. Ancillary to this was the critical question of whether genetically engineered microorganisms like HK44 could even survive under conventional environmental pressures for this length of time due to the extra metabolic burdens and lack of fitness that these recombinant microorganisms perceivably bear. Thus, HK44 populations were monitored during the initial two years of the release using selective plating and colony hybridization techniques, and it was shown that population concentrations decreased from an initial inoculum level of approximately 1 × 106 cfu/g soil to a final maintenance concentration of approximately 1 × 103 cfu/g soil. At these final low populations, however, HK44 populations remained in a physiological state conducive to reactivation and regrowth, and were still capable of generating bioluminescence when exposed to supplemental PAH contaminants and an inorganic nutrient amendment [1]. Based on this interesting outcome, it was decided that the lysimeter facility remain untouched and accessible for future longer-term population monitoring activities, and were thus covered with steel lids and remain today as a vegetation-free, desiccated soil ecosystem. In 2000, after an approximate four year residency, soils were again sampled and HK44 microbes and recombinant genes were yet again shown to be functionally active based on in situ bioluminescence emission and recovery of lux mRNA from the soils. With molecular experimental methods greatly advancing over the next decade, and with a newly sequenced HK44 genome in hand [5], it was decided in 2010 to perform another sampling to test the hypothesis that a recombinant microorganism (or its identifying genes) would still persist in the soil environment 14 years after its initial release. It was also anticipated that data collected from this sampling event would provide information on whether the bacterial decay constants identified in the original two year study would hold constant for the remaining twelve years. Additionally, a metagenomic analysis of the soil was performed to provide insight into the existing microbial community structure and how it may have been influenced by exposure to a recombinant population and/or chemical exposure.
Methods
Lysimeter field site
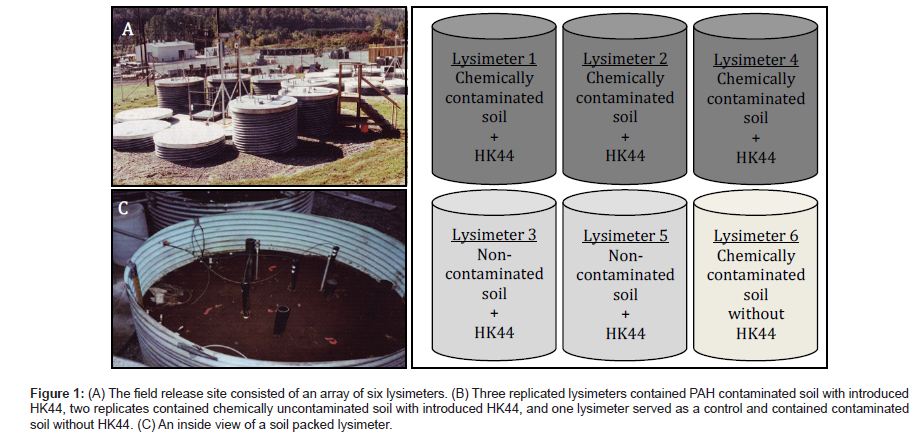
The field release experiments occurred within an array of six 4 m deep × 2.5 m diameter soil lysimeters constructed of epoxy-coated galvanized steel cylinders (Figure 1). Each lysimeter was filled with a stratified bed containing a bottom 31 cm layer of graded gravel, followed by a 61 cm layer of coarse sand, a 92 cm layer of clean soil, a 92 cm soil layer designated as the treatment zone, and a top layer of 61 cm of clean soil. Soils were of a Huntington loam type (42% sand, 40% silt, 18% clay, and 1.3% organic carbon) and were not presterilized before use and maintained their normal microbial biomass. The treatment layers received PAH-contaminated soil and/or an HK44 (DSM 6700) inoculum. Three lysimeters received a 92 cm layer of PAH-contaminated soil spray inoculated with strain HK44 (lysimeters 1, 2, and 4), two lysimeters received uncontaminated soil spray inoculated with strain HK44 (lysimeters 3 and 5), and one lysimeter served as a control and received a layer of PAH-contaminated soil not inoculated with strain HK44 (lysimeter 6). Each lysimeter was covered with a stainless steel lid that was removed only during sampling, thus preventing vegetation growth and moisture accumulation from rain. On day 135, lysimeters 1, 2, 4, and 6 each received a supplementary addition of chemical contaminants as a mixture of naphthalene and anthracene dissolved in Exxon Univolt transformer oil. Further specific details of the field release can be found in Ripp et al. [1], Ford et al. [4], and Cox et al. [6].
Figure 1: (A) The field release site consisted of an array of six lysimeters. (B) Three replicated lysimeters contained PAH contaminated soil with introduced HK44, two replicates contained chemically uncontaminated soil with introduced HK44, and one lysimeter served as a control and contained contaminated soil without HK44. (C) An inside view of a soil packed lysimeter.
Soil sampling and bacterial cultivation
Soil samples were collected from each lysimeter using a 2.3 cm diameter coring device that penetrated from the top soil layer, through the treatment layer, and into the soil layer beneath to capture a 1.8 m length transect. The top 61 cm of clean soil was discarded. The remainder of the core transects were then divided into four 30.5 cm subsamples. Six soil cores were removed from each lysimeter during the latest 2010 sampling. Over the lifetime of lysimeter sampling events, soil cores removed from each lysimeter ranged from 4 to 12.
Colony counts from each soil core subsample were obtained by resuspending 1 g of soil in 9 ml of sterile 0.1% sodium pyrophosphate and vortexing for 1 min. Ten-fold serial dilutions were then made in sterile phosphate buffered saline (PBS; in g/L, NaCl, 8; KCl, 0.2; Na2HPO4, 1.15; KH2PO4, 0.2) and 0.1 ml of the dilutions were plated in triplicate on yeast extract-peptone-glucose plates (YEPG; in g/L, yeast extract, 0.2; polypeptone, 2.0; glucose, 1.0; NH4NO3, 0.2) to obtain heterotrophic counts and on yeast extract-peptonesodium salicylate plates (YEPSS; in g/L, yeast extract, 0.2; polypeptone, 1; sodium salicylate, 0.5; sodium succinate, 2.7; NH4NO3, 0.2) supplemented with tetracycline at 14 mg/L to select for HK44 and other tetracycline resistant microbes. All plates were also supplemented with cycloheximide at 14 mg/L to inhibit fungal growth. Plates were incubated in the dark at room temperature and colonies were counted after 4 days. Since the YEPSS + tetracycline plates were not exclusively selective for P. fluorescence HK44, each of these plates was scanned for bioluminescent colonies using a PerkinElmer/Caliper Life Sciences IVIS Lumina photon counting camera system.
Given that P. fluorescens is known to exist in a viable-butnonculturable (VBNC) state, and that the microorganisms in the lysimeters are after a 14 year residency most likely in a nutrient starved state, additional resuscitation cultures were implemented to assist in reviving potential VBNC HK44 cells. Soil inoculums were introduced into 1:10 and 1:100 strength YEPG and minimal salts media (MSM; in g/L, KH2PO4, 0.68; K2HPO4, 1.73; MgSO4∙7H2O, 0.1; NH4NO3, 1.0; pH 7.0) supplemented with Exxon Univolt transformer oil, naphthalene, or a combination of both at concentrations of 10 and 100 ppm. Cultures were incubated in the dark with shaking at room temperature and sampled as described above at weekly intervals over an approximate six month period.
Quantitative PCR
DNA was extracted from 0.5 g of soil obtained from 18 subsamples from each of lysimeters 1, 2 and 4 using the Fast DNA Spin Kit for soil (QBiogene/MP Biomedicals, Solon, Ohio, USA). The DNA was diluted to 5 ng/μl before quantitative PCR analyses. Primers and probes were designed to target the nahA, tetA, and luxA genes found on the pUTK21 plasmid of strain HK44 (GenBank accession numbers AFOY01000001-AFOY01000130) (Table 1). Quantitative PCR (qPCR) reactions were prepared and run following previously published protocols that included duplicate six point standard curves generated from HK44 genomic DNA and spike controls for each sample [7,8]. For each assay, duplicate wells for six individual DNA extracts from each lysimeter were tested (total of 36 DNA extracts). Positives were determined as having at least 1 gene copy in each of the two duplicate wells. For positive samples, gene copies/g soil were calculated from the copies/well based on the DNA dilution factor and the amount of DNA extract per g of soil.
| Assay Gene Target (annealing temperature) | Oligonucleotide Sequencesa |
|---|---|
| nahA (60°C) |
HK44NahAc182f: 5'-tttgtttgcagctatcacgg-3' HK44NahAc302r: 5'-gcaaccgtagatgaagccat-3' HK44NahAc263Taqr: 5'-(Fam)cgagcgacttctttcaaccccagac(BHQ1)-3' |
| tetA (60°C) |
HK44TetA1055f: 5'-AGGTGGATGAGGAACGTCAG-3' HK44TetA1152r: 5'-ATAGATCGCCGTGAAGAGGA-3' HK44TetA1102Taqr: 5'-(Fam)GACGATCGAGGTCAGGCTGGTGAG(BHQ1)-3' |
| luxA (60°C) |
HK44LuxA29f: 5'-ATCAACCACCAGGTGAAACTCA-3' HK44LuxA167r: 5'-TCCCGTAAGACCAAACTCTGT-3' HK44LuxATaq54f: 5'-(Fam)AAGTAATGGATCGCTTTGTTCGGCTTGGT(BHQ1)-3' |
| a Present study. Probes were synthesized with FAM (fluorescein) and black hole quencher 1 (BHQ1) from Biosearch Technologies (Novato, CA, USA). | |
Table 1: Quantitative PCR primer and probes used to target specific genes carried on the P. fluorescens HK44 plasmid pUTK21.
Metagenomic analysis of soil samples
Phylogenetic analysis of soil samples was performed using multiplex tag-encoded pyrosequencing [9,10]. Due to the costs involved, only a pooled sampling of DNA from lysimeter 2 was used. DNA extracts were originally obtained in 2010 and then again one year later in 2011 on lysimeter 2 soil samples that had been archived under refrigeration at 4°C. To generate amplicons for pyrosequencing, 100 ng of each sample DNA was amplified using the eubacterial primers 27f and 1492r [11]. A second PCR was performed on each PCR product using barcoded fusion primers containing a unique 8 bp code [12] and targeting the V3 region of the 16S rRNA gene [13]. Emulsion PCR (emPCR) was performed and the amplicons were sequenced using both the A and B sequencing primers of the Roche 454 titanium chemistry. Following sequencing and data processing to base calls, the combined sequences in a FASTA file were parsed into individual samples using the Ribosomal Database pyrosequencing pipeline [14]. Individual sample sequences were then uploaded to MG-RAST for phylogenetic identification and analyses. The 2011 amplicon library (4469439.3) had 2,155 sequences after quality control (QC) and the 2010 amplicon library (4469438.3) had 6,975 sequences after QC.
Shotgun metagenomic DNA libraries were similarly prepared from pooled DNA extracts from original and 4°C stored lysimeter 2 soil samples. The rapid DNA library prep, emPCR, and sequencing using titanium chemistry were performed following Roche 454 protocols. Following sequencing and data processing to base calls, the signal flowgram files (sff) were uploaded to MG-RAST for annotation and analyses [14]. After QC, the 2010 DNA library (MG-RAST id 4466990.3) had 28,679,468 bp with 54,405 predicted ORFs and 24,172 RNA features. The 2011 DNA library from soil stored at 4°C (MGRAST id 4466989.3) had 15,767,619 bp with 30,862 predicted ORFs and 13,398 RNA features.
Results and Discussion
Quantification of soil heterotrophs and tetracycline resistant bacteria 14 years after inoculation with P. fluorescens HK44
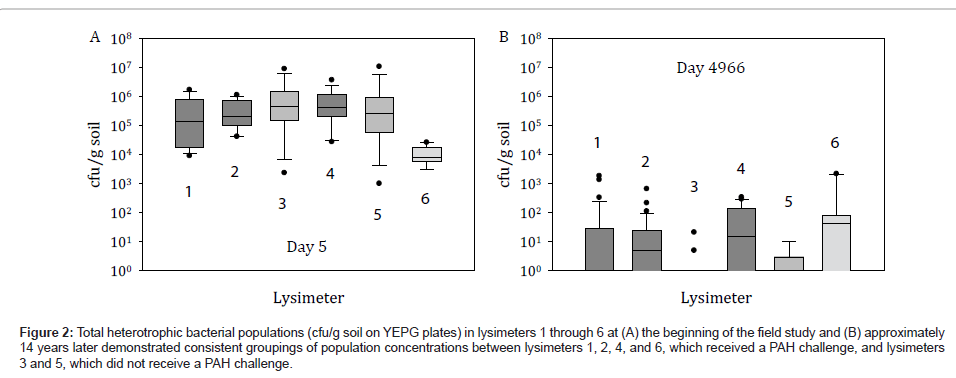
Although the environmental release of HK44 was conducted under controlled conditions in closed steel containers that restrict the cycling of carbon, nitrogen, phosphorus and water and the activities of other life forms, the lysimeters provided a replicated experimental design with undisturbed soils relevant to studying the relationships of soil properties on community structure and complexity. Furthermore, the lysimeters are more indicative of actual field performance and ecological consequence than that achievable in conventional laboratory microcosms. Comparison of total heterotrophic bacteria cultured on YEPG at the beginning of the lysimeter experiment and after 14 years indicates that heterotrophs decreased 100 to 1000-fold during this time (Figure 2). Our data also suggests that these heterotrophic microbial populations were established at the beginning of the lysimeter study and were based on PAH and oil content, evidently from the similar bacterial concentrations that persisted among PAH challenged lysimeters 1, 2, 4, and 6 and the analogously similar concentrations that persisted among the non-PAH challenged lysimeters 3 and 5 (Figure 2). Although PAH concentrations were highly variable in the soils due to uneven distribution, in general after 474 days PAHs remained at minimal levels ranging from 10 ppm to below detection limits [1]. Therefore, for the past approximate 12.5 years there has been little to no selective pressure in regards to PAH contaminants.
Figure 2: Total heterotrophic bacterial populations (cfu/g soil on YEPG plates) in lysimeters 1 through 6 at (A) the beginning of the field study and (B) approximately 14 years later demonstrated consistent groupings of population concentrations between lysimeters 1, 2, 4, and 6, which received a PAH challenge, and lysimeters 3 and 5, which did not receive a PAH challenge.
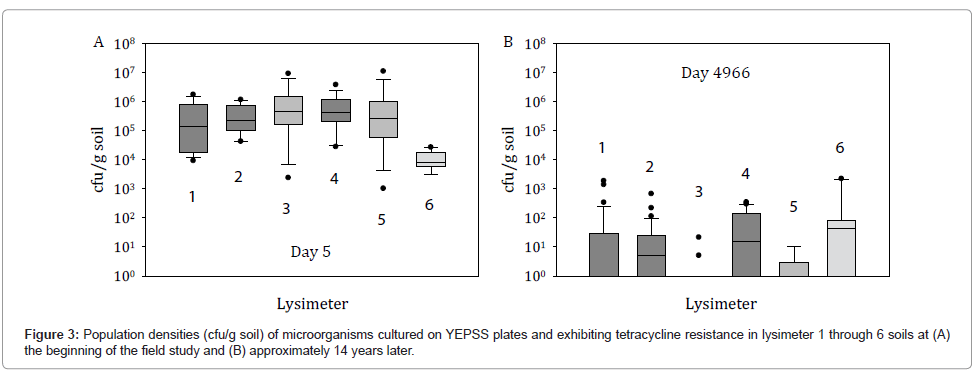
After 14 years, HK44-like bacteria were not recovered by selective plating on YEPSS supplemented with tetracycline. The small number of colonies obtained, typically at concentrations <100 cfu/g soil, although tetracycline resistant, did not display a bioluminescent phenotype when photon imaged (Figure 3). Attempts at recovering VBNC HK44 populations were similarly unproductive. This suggests several possibilities with respect to the presence or absence of strain HK44 and the ability of the recombinant genes to transfer to other hosts. First, strain HK44 may be present in the soils at concentrations below the detection limits of standard plate count methods and/or other microorganisms better able to grow on the selective media outcompete strain HK44 because it exists probably in a VBNC state. Thus, with the appropriate application of complex enrichment methods, it may be possible to resuscitate HK44 microbes. Those lengthy experiments are ongoing. Second, the Pseudomonas host containing the recombinant plasmid pUTK21 may have disappeared but all or part of this plasmid,such as the transposon element containing the lux and tetracycline resistance genes, may have transferred to a member or members of the indigenous microbiota, or persists in the soil as free DNA. Using the genomic approaches described below, we attempted to identify whether nucleic acid signature sequences from strain HK44 or its plasmid could be detected within the lysimeter soils.
Modeling the 14 year decline of the lysimeter microbial populations
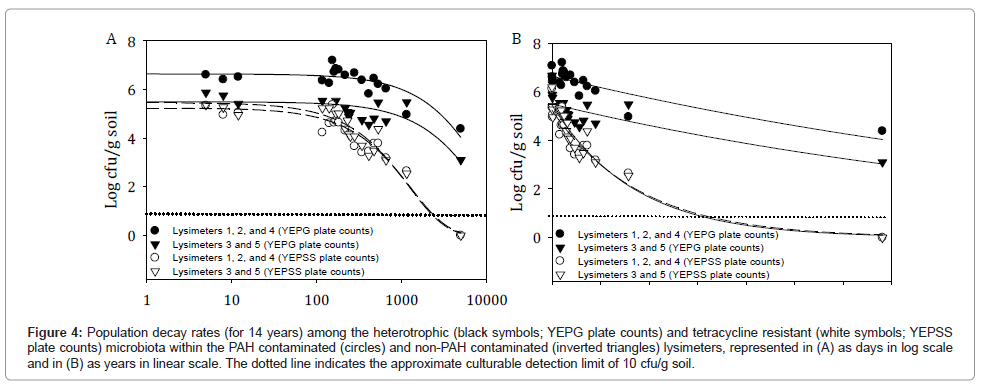
Figure 4 depicts the 14 year 2 parameter exponential decay curves for combined lysimeters 1, 2, and 4 (PAH challenged) and combined lysimeters 3 and 5 (not PAH challenged) for both the total heterotrophic bacterial populations (YEPG plate counts) and the tetracycline resistant bacteria populations inclusive of strain HK44 (YEPSS plate counts). These curves predict minimal differences in the decline of tetracycline resistant HK44-like populations to below detection limits between the PAH contaminated and non-contaminated lysimeters, with similar decay rates of 0.0008 log cfu/day. Lysimeters 1, 2, and 4 reach this milestone at approximately 1,782 days (4.9 years) and lysimeters 3 and 5 at approximately 1,843 days (5.0 years) (Table 2). This was anticipated based on the removal of the selective PAH pressures as early as 474 days into the experiment.
Figure 4: Population decay rates (for 14 years) among the heterotrophic (black symbols; YEPG plate counts) and tetracycline resistant (white symbols; YEPSS plate counts) microbiota within the PAH contaminated (circles) and non-PAH contaminated (inverted triangles) lysimeters, represented in (A) as days in log scale and in (B) as years in linear scale. The dotted line indicates the approximate culturable detection limit of 10 cfu/g soil.
| Treatment | Initial log concentration (T=0) (a) | Decay rate (-b) in days | Decay rate (-b) in years | Adjusted r2 | Predicted time to decline below detection limit |
|---|---|---|---|---|---|
| Lysimeters 1, 2, and 4; YEPG plate counts | 6.6430 | -0.0001 | -0.0366 | 0.6849 (P < 0.0001) |
16,630 days (45.6 years) |
| Lysimeters 1, 2, and 4; YEPSS plate counts | 5.2432 | -0.0008 | -0.2801 | 0.9013 (P < 0.0001) |
1,782 days (4.9 years) |
| Lysimeters 3 and 5; YEPG plate counts | 5.4976 | -0.0001 | -0.0438 | 0.5677 (P = 0.0003) |
14,740 days (40.4 years) |
| Lysimeters 3 and 5; YEPSS plate counts | 5.4616 | -0.0008 | -0.2973 | 0.9056 (P < 0.0001) |
1,843 days (5.0 years) |
Table 2: Decay rate data for a 2 parameter exponential decay curve for log cfu/g soil versus log time (days) or time (years) using the equation y = ae-bx.
For total heterotrophic populations, PAH contaminated lysimeters 1, 2, and 4 are predicted to be maintained at detectable levels for 16,630 days (45.6 years) as opposed to uncontaminated lysimeters 3 and 5 remaining detectable for 14,740 days (40.4 years). The extended survival of heterotrophic populations in the PAH contaminated lysimeters was most likely due to the spike in bacterial concentrations that occurred after the addition of supplementary contaminants on day 135 (increasing from 1.8 × 106 cfu/g soil on day 141 to 1.6 × 107 cfu/g soil on day 154 [1]) as the calculated decay for both types of lysimeter is the same at 0.0001 log cfu/day.
Quantitative PCR
The HK44-specific luciferase alpha chain (luxA) and tetracycline resistance (tetA) genes were identified by qPCR at low concentrations (~100 to 5000 copies/g soil). The luxA gene was identified in one lysimeter 2 DNA sample. The tetA gene was identified in one lysimeter 1 and one lysimeter 2 DNA sample (Table 3). No nahA gene signatures were identified in any of the samples. The luxA and tetA products were purified, cloned, and sequenced. The luxA clone (131 bp) had a 100% similarity match to the luxA gene sequence of strain HK44 and a 99% similarity match to the Vibrio fischeri MJ11 chromosome II (GenBank id CP001133.1) alkanal monooxygenase alpha chain (bacterial luciferase alpha chain). The tetA clones (98 bp each) had a 100% similarity match to the tetA gene sequence of strain HK44 and a 100% similarity match to the Escherichia coli strain A21 tetracycline resistance protein gene (GenBank id HQ33263.1). These results suggest that although HK44 was no longer culturable from the soil lysimeters, HK44 specific gene remnants were present at very low but still detectable concentrations.
| Lysimeter | Conditions | Number of samples | Heterotrophic cfu/g (geomean) | YEPSS phenotypea cfu/g (geomean; % of positive samples) | Quantitative PCR copies/g (% of positive samples) | ||
|---|---|---|---|---|---|---|---|
| nahA | tetA | luxA | |||||
| 1 | PAH contaminated + HK44 | 36 | 8904 | 78 (33%) | NDb | 4455 (17%) | ND |
| 2 | PAH contaminated + HK44 | 36 | 14910 | 18 (61%) | ND | 872 (17%) | 2052 (17%) |
| 3 | Not contaminated + HK44 | 36 | 870 | 10 (6%) | ND | ND | ND |
| 4 | PAH contaminated + HK44 | 20 | 23924 | 49 (13%) | ND | ND | ND |
| 5 | Not contaminated + HK44 | 21 | 957 | 7 (19%) | ND | ND | ND |
| 6 | PAH contaminated without HK44 | 14 | 6189 | 65 (43%) | ND | ND | ND |
Table 3: Summary of measured parameters of soils sampled from six lysimeters after 14 years of inoculation with PAHs and P. fluorescens HK44.
Metagenomic analyses
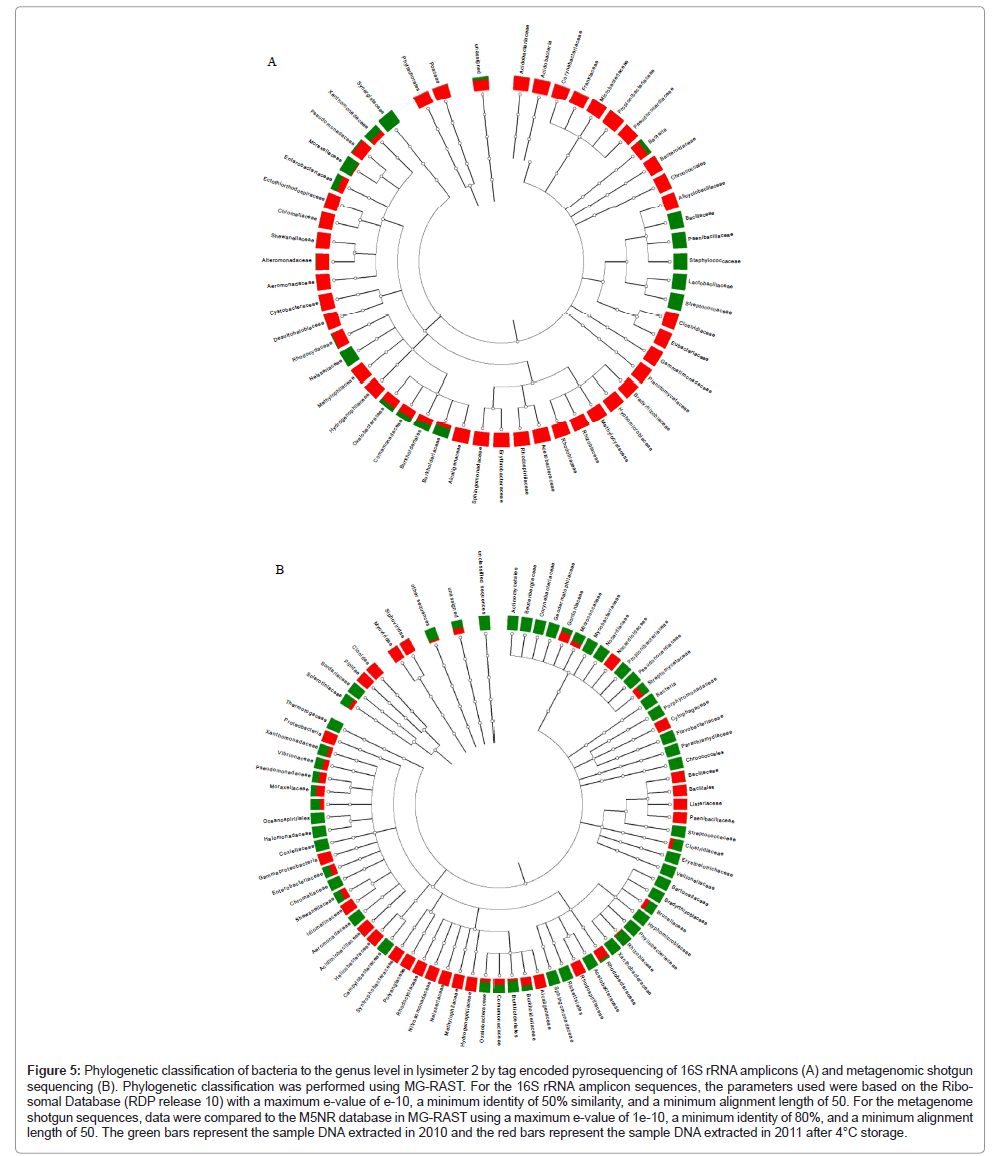
Based on the quantitative PCR results, pooled DNA extracts from lysimeter 2 were chosen for further phylogenetic and metagenomic analyses to determine whether P. fluorescens bacteria and/or specific HK44-like sequences could be detected. Based on 16S rRNA genes, the dominant bacterial genus found in lysimeter 2 directly after sampling was Burkholderia (95% of sequences) with Pseudomonas sequences found at <0.1% (Figure 5, Table 4). Interestingly, after storing the lysimeter soils for one year at 4°C in the DNA extracted from the stored samples, the Burkholderia component decreased to 2.6% while the Pseudomonas component increased to 18.7%. The phylogenetic composition of the 2010 versus 2011 DNA extracts by metagenomic analyses also indicated that Burkholderia was more abundant in the sample extracted before storage at 4°C than after storage at 4°C (79% and 5.6% Burkholderia sequences, respectively). Slightly higher Pseudomonas sequences were detected in the DNA extracted from the sample stored at 4°C than the DNA extracted before storage at 4°C at a similar concentration (3.8% and 2.1%, respectively). The α-diversity, which is a measure of microbial species diversity, was 51.7 species for the sample stored at 4°C and 7.4 species for the un-stored sample. This indicates that the sample stored at 4°C had ~10-fold more species diversity than the sample initially collected. The lower species complexity in the initial sample likely reflects the dominance of the Burkholderia species that was subsequently reduced under storage at 4°C. Five sequences were annotated in MG-RAST as P. fluorescens, four from the samples stored at 4°C and one from the sample before storage. One sequence was 99% similar to HK44 as determined by blasting the sequences to HK44 sequences stored in RAST [15]. This one sequence represented approximately 0.05% of the sequence population.
Figure 5: Phylogenetic classification of bacteria to the genus level in lysimeter 2 by tag encoded pyrosequencing of 16S rRNA amplicons (A) and metagenomic shotgun sequencing (B). Phylogenetic classification was performed using MG-RAST. For the 16S rRNA amplicon sequences, the parameters used were based on the Ribo-somal Database (RDP release 10) with a maximum e-value of e-10, a minimum identity of 50% similarity, and a minimum alignment length of 50. For the metagenome shotgun sequences, data were compared to the M5NR database in MG-RAST using a maximum e-value of 1e-10, a minimum identity of 80%, and a minimum alignment length of 50. The green bars represent the sample DNA extracted in 2010 and the red bars represent the sample DNA extracted in 2011 after 4°C storage.
| MG-RAST ID | Amplicon samples | # ribosomal sequences (RDPa) | # Burkholderia genus (% of total) | # Pseudomonas genus (% of total) |
|---|---|---|---|---|
| 4469438.3 | Lysimeter 2, 2010 amplicons obtained directly after sampling | 6815 | 6467 (95%) | 5 (<0.1%) |
| 4469439.3 | Lysimeter 2, 2011 amplicons obtained one year after storage at 4°C | 2155 | 42 (2.6%) | 299 (18.7%) |
| MG-RAST ID | Metagenome samples | # of bacterial taxonomic hits | # Burkholderia genus (% of total) | # Pseudomonas genus (% of total) |
| 4466990.3 | Lysimeter 2, 2010 metagenome obtained directly after sampling | 97914 | 77437 (79%) | 2060 (2.1%) |
| 4466989.2 | Lysimeter 2, 2011 metagenome obtained one year after storage at 4°C | 36960 | 2060 (5.6%) | 1417 (3.8%) |
| aRDP, Ribosomal Database Project annotation | ||||
Table 4: Distribution of Burkholderia and Pseudomonas in soil of lysimeter number 2, analyzed using DNA extraction from soil sampled in 2010 and after storage for one year at 4°C as determined by tag encoded pyrosequencing and metagenomic analyses.
Metagenomic DNA libraries were made from the same samples used to generate pyrosequencing amplicon libraries and annotated and analyzed in MG-RAST. Again, the calculated α-diversity of the sample stored at 4°C (564.9 species) was much larger than the original, unstored samples (42.9 species). Several searching methods were used to identify potential HK44 sequences based on phylogenetic classification of the sequences. In the first method, 55 sequences identified as P. fluorescens Pf0-1 using GenBank in MG-RAST in the sample stored at 4°C were downloaded and blasted against the HK44 sequence in RAST. The highest sequence match was 96% for a ribosomal protein but this sequence was not considered diagnostic of HK44. Similarly, 73 sequences were identified as P. fluorescens Pf0-1 using GenBank in MGRAST in the sample before storage at 4°C. None of these samples had matches greater than 95% with HK44 sequences. Three sequences were identified as belonging to the P. fluorescens PC20 plasmid pNAH20 (nahH, nahO, and nahN). However, when they were compared to HK44 sequences they had a less than 95% sequence match suggesting that they were not likely to be identifiable with the pUTK21 plasmid. Sequences for naphthalene and biphenyl degradation genes were identified in MG-RAST using functional annotation analyses (Table 5). In addition to the three lower pathway genes described above (nahH, nahO, and nahN), three upper pathway naphthalene dioxygenase genes were identified. These three genes were not related to those found in strain HK44 but rather were similar to naphthalene dioxygenases found in Rhodococcus, Burkholderia, and Mycobacterium species. Multiple biphenyl degrading genes were also found. The genes identified were almost exclusively found in Burkholderia, which is consistent with the dominance of Burkholderia sequences found in the shotgun library metagenome and amplicon sequences. The identification of these genes for naphthalene and biphenyl degradation was also of interest because the original soil used to fill these lysimeters was uncontaminated soil, suggesting that these genes may be widely distributed in soils [16-18].
| Gene name | MG-RAST annotation | GenBank organism (e value, % sequence similarity) |
|---|---|---|
| nahH gene for catechol 2,3-dioxygenase | gb|AAW81680.2| catechol 2,3-dioxygenase [Pseudomonas fluorescens] | dbj|AB266142.2| Uncultured bacterium DNA, fosmid clone, clone: 09B01 (1e-142, 88%) emb|AM406670.1| Azoarcus sp. BH72, complete genome (5e-52, 72%) |
| nahO (acetaldehyde dehydrogenase) | gb|AAA89106.1| acetaldehyde dehydrogenase (acylating) [Pseudomonas putida] | gb|CP000152.1| Burkholderia sp. 383, complete sequence (0.0, 98%) gb|AF491307.2| Pseudomonas putida NCIB 9816-4 plasmid pDTG1, complete sequence (7e-109, 78%) |
| nahN (2-hydroxymuconic semialdehyde hydrolase) | gb|ACQ63519.1 4|-hydroxy-2-ketovalerate aldolase [Pseudomonas fluorescens] | gb|AY887963.3| Pseudomonas fluorescens strain PC20 plasmid pNAH20, complete sequence (1e-98, 78%) |
| Naphthalene dioxygenase | gb|AAD13057.1| naphthalene dioxygenase [uncultured bacterium U2a] | gb|CP002329.1|Mycobacterium sp. JDM601 (5e-22, 84%) |
| Naphthalene 1,2-dioxygenase system ferredoxin component | gb|ABC35912.1| naphthalene 1,2-dioxygenase system ferredoxin component [Burkholderia thailandensis E264] | gb|CP000459.1| Burkholderia cenocepacia HI2424 chromosome 2 (0.0, 100%) |
| 1,2-dihydroxynaphthalene dioxygenase | gb|ACL31223.1| 1,2-dihydroxynaphthalene dioxygenase [Rhodococcus sp. TFB] | gb|AJ937590.1| Uncultured bacterium partial bphC gene for extradiol dioxygenase, clone B04 (9e-177, 92%) gb|CP000676.1| Novosphingobium aromaticivorans DSM 12444 plasmid pNL1, complete sequence (2e-57, 75%) |
| Biphenyl dioxygenase large subunit | gb|ACV31375.1| biphenyl dioxygenase large subunit [Sphingomonas sp. DN1] | dbj|AP010947.1| Azospirillum sp. B510 plasmid pAB510a DNA (2e-22, 67%) |
| 2,3-dihydroxybiphenyl-1,2-dioxygenase | gb|AAF04139.1| 2,3-dihydroxybiphenyl-1,2-dioxygenase [Pseudomonas sp. SY5] | gb |AM747721.1| Burkholderia cenocepacia J2315 chromosome 2, complete genome (0.0, 95%) |
| Biphenyl-2,3-diol 1,2-dioxygenase (23ohbp oxygenase) (2,3-dihydroxybiphenyl dioxygenase) (dhbd) | gb|EEE08827.1| biphenyl-2,3-diol 1,2-dioxygenase (23ohbp oxygenase) (2,3-dihydroxybiphenyl dioxygenase) (dhbd) [Burkholderia multivorans CGD2] | gb|CP000699.1| Sphingomonas wittichii RW1, complete genome (5e-21, 67%) |
| cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase (bphB) | gb|ABE37054.1| Cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase (BphB) [Burkholderia xenovorans LB400] | gb|AF061751| Burkholderia sp. strain RP007 PAH-catabolic gene cluster (3e-24, 76%) |
| Oxido-reductase/dehydratase | gb|AAO64287.1| putative oxidoreductase [Pseudomonas putida] | gb|CP002599.1| Burkholderia gladioli BSR3 chromosome 1, complete sequence (1e-41, 70%) |
Table 5: Biodegradative sequences for naphthalene and biphenyl genes identified in MG-RAST functionally annotated by GenBank (e-10, 50%similarity, 50% minimum sequence alignment).
Conclusion
The introduction of genetically engineered bacterial strains into previously characterized microbial communities allows us to decipher fundamental questions regarding the interactions among microbes that enhance or diminish their persistence and their potential risk. At the time of the field release, now over 15 years ago, it was generally assumed that engineered microorganisms would be unable to effectively compete with the indigenous microbial consortia because they had a reduced level of fitness due to the extra energy demands mandated by the presence of the introduced genes and by the fact that they were extensively nurtured under optimal laboratory conditions,thus making them physiologically less fit and less prone to survive. Additionally, recombinant microbes designed to survive under certain selective pressures were expected to die off as those selective pressures were removed from their environment. Models at the time, in fact, predicted that engineered microorganisms would generally survive for three years in their introduced environment [19]. The lysimeter study clearly provides evidence to the contrary, and provides a one-of-a-kind risk assessment paradigm for understanding the molecular mechanisms involved in the environmental dissemination of recombinant genes. The HK44 microbial population could be revived from the lysimeter soils four years after its release, even though the selective pressure of the hydrocarbon contaminants of which it degrades was depleted at least two years prior [1]. Importantly, the inability to culture microorganisms does not mean that they have disappeared from the microbial community, as demonstrated here where signature genetic elements related to strain HK44 were identified 14 years after their initial soil introduction. Given that soil microbial communities have thousands of individual species representing population levels estimated at ~5 × 1030 cells, it is quite possible that soil microbes comprising <1% of the total community, the approximate limit of detection by culturable methods, can still remain important and influential members of a community [20]. Our results suggest that our ability to predict survivability of particular microorganisms based on a limited number of genetic markers is poor, and more studies are needed to understand inter- and intra-species competition at the genomic, metabolic, and functional levels, especially from a risk assessment perspective. Based on ecological theories, it is intuitive that competition must exist between microbes, particularly when nutritional substrates are limiting. Even with recent advances in omics, our ability to differentiate between species is limited due to the predominant complexity of soil microbial systems. Thus, studying a uniquely introduced microorganism like P. fluorescens HK44 offers a unique opportunity to examine the metagenomic impact and potential environmental risk profile of a recombinant microbe over a markedly long term residency.
Acknowledgements
The authors would like to acknowledge the financial support of the USDA National Institute of Food and Agriculture Biotechnology Risk Assessment Program under grant number 2009-39210-20230. Sampling of the lysimeters was partially supported by the United States Department of Energy under the Environmental Remediation Sciences Program (ERSP), Office of Biological and Environmental Research, Office of Science. Oak Ridge National Laboratory is managed by University of Tennessee UT-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
References
- Ripp S, Nivens DE, Ahn Y, Werner C, Jarrell J, et al. (2000) Controlled field release of a bioluminescent genetically engineered microorganism for bioremediation process monitoring and control. Environ Sci Technol 34: 846-853.
- King JMH, DiGrazia PM, Applegate B, Burlage R, Sanseverino J, et al. (1990) Rapid, sensitive bioluminescence reporter technology for naphthalene exposure and biodegradation. Science 249: 778-781.
- Trogl J, Kuncova G, Kubicova L, Parik P, Halova J, et al. (2007) Response of the bioluminescent bioreporter Pseudomonas fluorescens HK44 to analogs of naphthalene and salicylic acid. Folia Microbiol 52: 3-14.
- Ford CZ, Sayler GS, Burlage RS (1999) Containment of a genetically engineered microorganism during a field bioremediation application. Appl Microbiol Biotechnol 51: 397-400.
- Chauhan A, Layton AC, Williams DE, Smartt AE, Ripp S, et al. (2011) Draft genome sequence of the polycyclic aromatic hydrocarbon-degrading, genetically engineered bioluminescent bioreporter Pseudomonas fluorescens HK44. J Bacteriol 193: 5009-5010.
- Cox CD, Nivens DE, Ripp S, Wong MM, Palumbo A, et al. (2000) An intermediate-scale lysimeter facility for subsurface bioremediation research. Bioremediation 4: 69-79.
- Layton A, McKay L, Williams D, Garrett V, Gentry R, et al. (2006) Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl Environ Microbiol 72: 4214-4224.
- Hawkins SA, Robinson KG, Layton AC, Sayler GS (2008) Response of Nitrobacter spp. ribosomal gene and transcript abundance following nitrite starvation and exposure to mechanistically distinct inhibitors. Environ Sci Technol 42: 901-907.
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, et al. (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A 103: 12115-12120.
- Acosta-Martinez V, Dowd S, Sun Y, Allen V (2008) Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol Biochem 40: 2762-2770.
- DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, et al. (2007) High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol 53: 371-383.
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R (2008) Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235-237.
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, et al. (2008) Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PloS Genet 4: 1000255.
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, et al. (2008) The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9: 386.
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. (2008) The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9: 75.
- Wang Q, Zhang S, Zou L, Xie S (2011) Impact of anthracene addition on microbial community structure in soil microcosms from contaminated and uncontaminated sites. Biomed Environ Sci 24: 543-549.
- Labbe D, Margesin R, Schinner F, Whyte LG, Greer CW (2007) Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated Alpine soils. FEMS Microbiol Ecol 59: 466-475.
- DeBruyn JM, Mead TJ, Wilhelm SW, Sayler GS (2009) PAH biodegradative genotypes in Lake Erie sediments: Evidence for broad geographical distribution of pyrene-degrading Mycobacteria. Environ Sci Technol 43: 3467-3473.
- Van der Hoeven N, van Elsas JD, Heijnen CE (1996) A model based on soil structural aspects describing the fate of genetically modified bacteria in soil. Ecol Model 89: 161-173.
- Vogel TM, Simonet P, Jansson JK, Hirsch PR, Tiedje JM, et al. (2009) TerraGenome: a consortium for the sequencing of a soil metagenome. Nat Rev Microbiol 7: 252.
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14546
- [From(publication date):
specialissue-2012 - Dec 03, 2024] - Breakdown by view type
- HTML page views : 10099
- PDF downloads : 4447