Research Article Open Access
Bioremoval of Azo dye Reactive Red by Bacillus spp. ETL-1982
| Maulin P Shah*, Patel KA, Nair SS and Darji AM | |
| Applied & Environmental Microbiology Lab, Enviro Technology Limited (CETP), Plot No: 2413/2414, GIDC, Ankleshwar- 393 002, Gujarat, India | |
| Corresponding Author : | Maulin P Shah Applied and Environmental Microbiology Lab Enviro Technology Limited (CETP), India Tel: +91-90 999 65504 Fax: +91- 2646-250707 E-mail: shahmp@uniphos.com |
| Received: March 11, 2013; Accepted: April 26, 2013; Published: April 28, 2013 | |
| Citation:Shah MP, Patel KA, Nair SS, Darji AM (2013) Bioremoval of Azo dye Reactive Red by Bacillus spp. ETL-1982. J Bioremed Biodeg 4:186. doi:10.4172/2155-6199.1000186 | |
| Copyright: © 2013 Shah MP, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Bacillus spp. ETL-1982, with notable ability to decolorize the reactive textile dye Reactive Red, was isolated from the activated sludge collected from a textile mill. Phenotypic characterization and phylogenetic analysis of the 16S rDNA sequence indicated that the bacterial strain belonged to the genus Bacillus. Bacillus ETL-1982 showed a strong ability to decolorize reactive textile dye at pH 9.0 and 37°C and with 4 gL-1 glucose concentrations was considered to be the optimum decolorizing conditions. Bacillus spp. ETL-1982 grew well in a high concentration of dye (300 mgl-1), resulting in approximately 96% decolorization extent in 36 h, and could tolerate up to 1000 mg l-1 of dye. UV–Vis analyses and colorless bacterial cells suggested that Bacillus spp. ETL-1982 exhibited decolorizing activity through biodegradation, rather than inactive surface adsorption. High decolorization extent and facile conditions show the potential for this Bacillus ETL-1982 to be used in the biological treatment of dyeing mill effluents.
| Keywords |
| Bacillus; Azo dye; Reactive black; Reactive yellow |
| Introduction |
| Reactive dyes, including many structurally different dyes, are extensively used in the textile industry because of their wide variety of color shades, high wet fastness profiles, ease of application, brilliant colors, and minimal energy consumption. The three most common groups are azo, anthraquinone and phthalocyanine [1]. Disposal of these dyes into the environment causes serious damage, since they may significantly affect the photosynthetic activity of hydrophytes by reducing light penetration [2]. They may be toxic to some aquatic organisms due to their breakdown products [3]. Dyes can be removed from wastewater by chemical and physical methods including adsorption, coagulation–flocculation, oxidation and electrochemical methods [4,5]. However, both the physical and chemical methods have many disadvantages in application, such as high-energy costs, highsludge production, and formation of by-products [6]. Conversely, bioprocessing can overcome these defects being cost effective and environmentally benign. Fungi [7-9] and algae [10,11] have been used in dye decolorization. Adsorption rather than degradation plays a major role during the decolorization process by fungi and algae, as a result, the dyes remain in the environment. It is well known that bacteria can degrade and even completely mineralize many reactive dyes under certain conditions [12-14]. Some new bacterial strains capable of decolorizing a broad-spectrum of dyes have also been isolated and characterized [15]. Bacterial degradation of reactive dyes is often initiated under anaerobic conditions by an enzymatic biotransformation step [16,17]. The resulting products such as aromatic amines are further degraded by multiple-step bioconversion occurring aerobically or anaerobically [18-20]). In this study, a bacterial strain Bacillus spp. ETL-1982, capable of decolorizing azo dyes was isolated and, in addition, the effects of various parameters (such as oxygen, initial glucose concentration, dye concentration, pH and temperature) on dye decolorization by the bacterial strain were investigated. It is noticeably found that Ankleshwar Industrial Zone is a Hub of Chemical and Textile industry, which requires appropriate attention in terms of pollution control. It is quite necessary to reduce the pollution load discharged by the textile industry. In connection to observe the major Environmental problems faced by the Industrial estate, a remarkable and affordable research task has been taken as a priority model to solve the problems of Industry of Ankleshwar area. |
| Materials and Methods |
| Chemicals & reagents |
| Reactive Red, a commonly used commercial reactive dye, was chosen for the screening of degradative bacteria. In addition, other reactive dyes such as Reactive Black, Reactive Yellow, and Reactive Blue were used to detect the decolorizing ability of strain ETL-1982 against structurally different dyes. |
| Isolation and identification |
| The dye-decolorizing bacterial strain was isolated from activatedsludge samples collected from wastewater treatment plants of dyeing industries in Ankleshwar, Gujarat, India. Screening of the strains for dye decolorization was performed by enrichment culture technique using NM9 and DNM9 media as described previously [21]. After purification by successive single colony isolation on a DNM9 agar plate, strain ETL-1982 was identified by carbon source utilization patterns using Biolog GN2 microplate (Biolog, USA) and the analysis of 16S rDNA sequences. For the 16S rDNA sequence analysis, bacterial genomic DNA was extracted and purified using a Wizard Genomic DNA Prep. Kit (Promega Corp., Madison). Two primers annealing to the 5` and 3` end of the 16S rRNA gene were 5`-GAGTTTGATCCTGGCTCAG-3` (positions 9 to 27 (Escherichia coli 16S rDNA numbering)) and 5`-AGAAAGGAGG TGATCCAGCC-3` (positions 1542 to 1525 (E. coli 16S rDNA numbering)), respectively. Polymerase Chain Reaction (PCR) was performed as follows: pre-denaturation at 95°C for 5 min, 30 cycles at 95°C for 40 s, 55°C for 40 s and 72°C for 2 min. The PCR product was subcloned into pGEM-T easy vector (Promega, Madison, USA) and its nucleotide sequence was determined by Banglore Genei Ltd. (Banglore, India). The partial rDNA sequences were analyzed using a BLAST search algorithm to estimate the degree of similarity to other rDNA sequences obtained from the NCBI/GenBank. Phylogenetic trees were constructed by the ClustalX program [22]. Physiological characteristics were determined according to the procedures outlined in Bergey`s Manual of Determinative Bacteriology [23]. |
| Effects of process parameters |
| The isolate Bacillus ETL-1982 was cultivated for 24 h in conical flasks containing 100 ml nutrient broth and was amended with 0.5 g l-1 of Reactive Red separately to determine the effect of varying pH on dye decolorization, ranging from 6.0 to 11.0. The dye decolorization was monitored at regular intervals. On similar lines, the optimum temperature of Bacillus ETL-1982 -mediated dye decolorization was determined by evaluating the dye decolorization at 20, 25, 30, 37, and 45°C. |
| Measurement of decolorization extent |
| Samples (0.4 mL) were collected every 12 h and centrifuged at 7000×g for 5 min. Decolorization extent was determined by measuring the absorbance of the culture supernatant at 560 nm using a Shimadzu- UV1800 visible spectrophotometer. Decolorization extent was calculated using the following equation: |
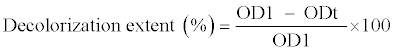 |
| Where OD1 refers to the initial absorbance, ODt refers to the absorbance after incubation; and t refers to the incubation time. |
| Decolorization study of Bacillus spp. ETL-1982 on reactive red at static & shaking condition |
| The isolate Bacillus ETL-1982 was cultivated for 24 h in conical flasks containing 100 ml nutrient broth and was amended separately with 0.5 g l-1 of Reactive Red. The dye degradation efficiency of Bacillus ETL-1982 was studied under static and shaking conditions (120 rpm) at 37°C in orbital shaking incubator (Remi CIS-24 BL). After different time intervals, aliquot (2 ml) of the culture media was withdrawn and centrifuged at 8000×g for 10 min in a refrigerated centrifuge (Remi, India) to separate the bacterial cell mass and supernatant. The supernatant is used for analysis of decolorization and all the experiments were repeated in triplicates. |
| Results and Discussion |
| Isolation and identification |
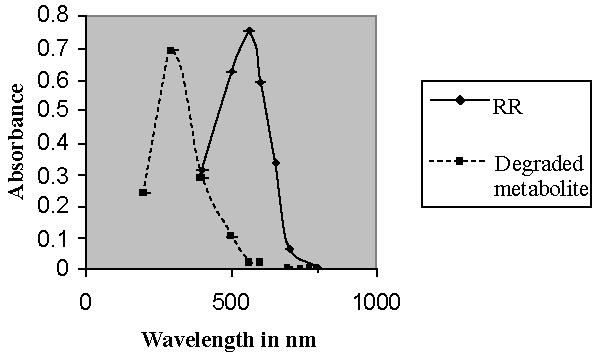
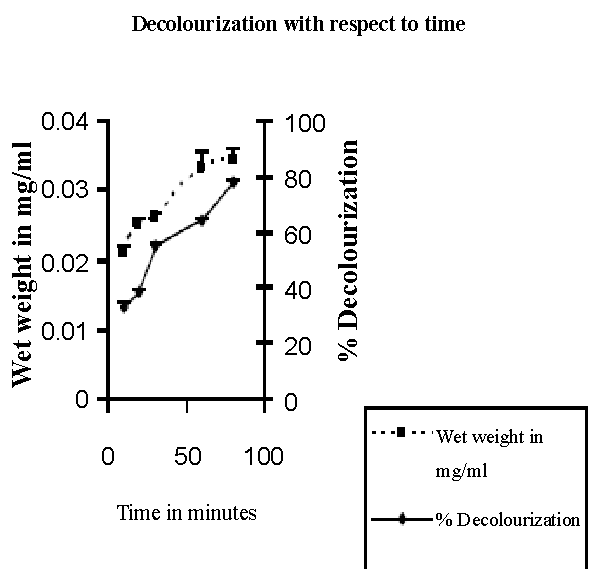
| Isolation of bacteria from mixed sample was carried out by the enrichment technique using nutrient broth and Reactive red as source of carbon and nitrogen. Decolorization occurred only when carbon and nitrogen sources were available for growth. An identification of the culture was based on 16sr-RNA analysis done by Bangalore Genei, Bangalore, India. The nucleotide alignment of this strain showed it was most phylogenetically similar to the Bacillus. Sequence analysis of 16S rDNA showed that strain ETL-1982 had highest similarity with the species Bacillus spp (96%) which has been proved to have decolorizing ability against azo dyes. Based on the phenotypic characteristics and phylogenetic analysis, strain ETL-1982 was identified as Bacillus (Figure 1). The absorbance peaks in the visible region disappeared indicating complete decolourization1. In the UV spectra the peak at 562 nm was replaced by new peak at 300nm (Figure 1a). Decolorization with respect to time showed within 80 minutes 77.73% decolorization along with simultaneous increase in wet weight of the cell. This indicated the ability of the isolate to grow in presence of the dye (Figure 1b). There was no abiotic loss of Reactive red within 24 hrs incubation indicating that the decolorization of Reactive red was due to biological mechanism rather than adsorption. To confirm whether this decolorization was due to the bacterial action or variation in pH, change in pH was recorded in the range of 7.4 ± 0.2. |
| Decolorization study of reactive red with reference to static and shaking condition |
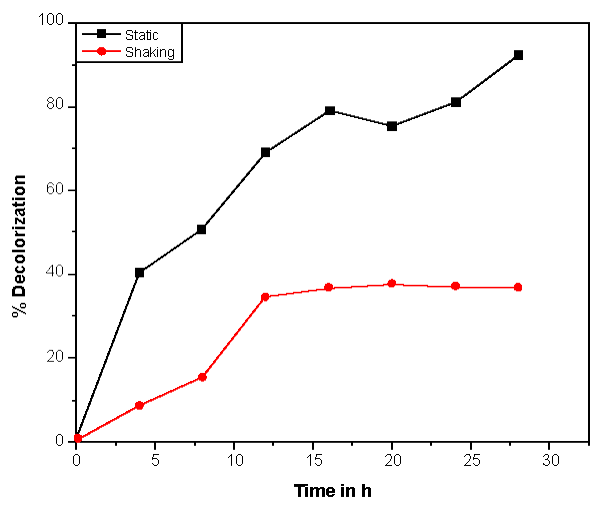
| Bacillus ETL-1982 was isolated and its decolorization potential was estimated. The decolorization was observed on UV visible spectrophotometer at different time interval. The UV-vis absorbance pattern of Reactive red showed complete degradation of dye (Figure 2). With appropriate calibrations at specific wavelengths, concentrations of dyes can be determined by using an UV–Visible spectrophotometer and the dye decolorization studies can be conducted and followed. The dye decolorization of azo dyes Reactive Red was studied under static and shaking conditions with an initial dye concentration of 0.5 g l-1. It was observed that under static conditions, the dye decolorization efficiency was 85-95% within 24 h as compared to 35% decolorization under shaking conditions (Figure 3). Anaerobic or static conditions were necessary for bacterial decolorization, though the cell growth was poorer than that under aerobic conditions. Therefore, static conditions were adopted to investigate bacterial decolorization in the following experiments. The results were similar to those of studies on Escherichia coli NO3 [24], Pseudomonas luteola [25] and Rhodopseudomonas palustris [26]. |
| Optimization of process parameters on decolorization of reactive red |
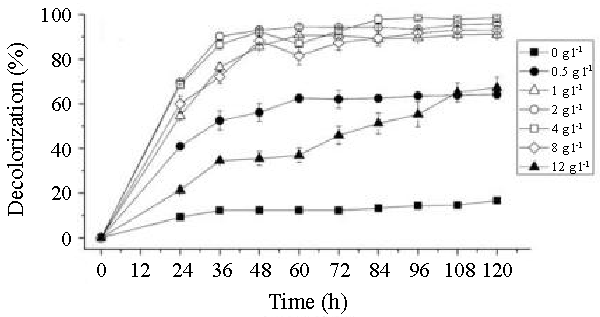
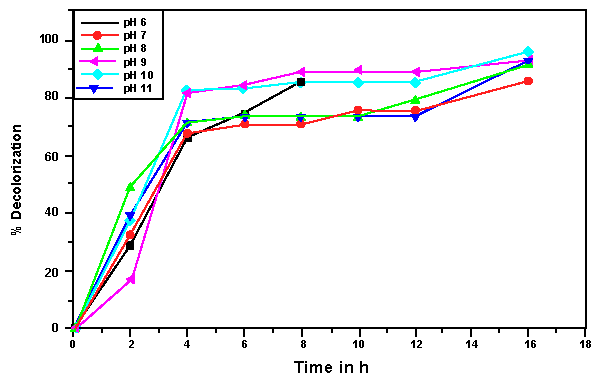
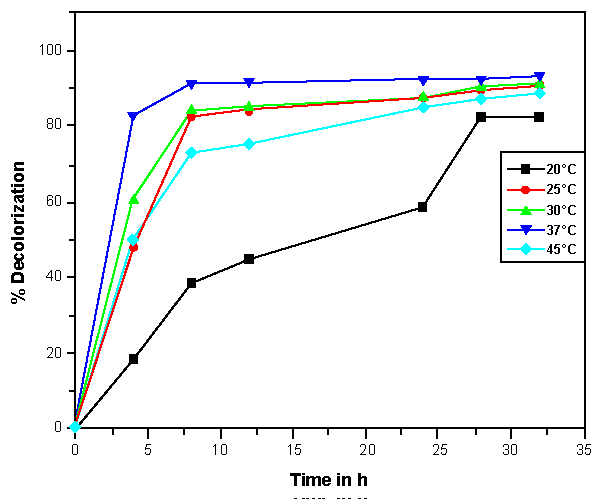
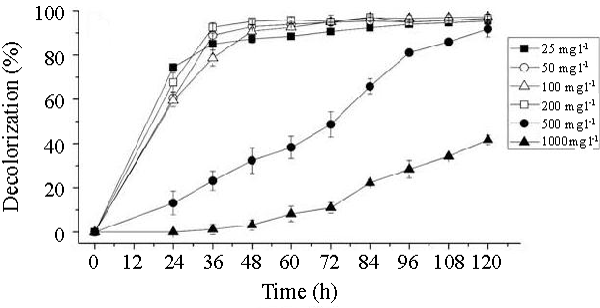
| The effects of initial glucose concentration, pH, temperature and initial dye concentration on decolorization by Bacillus spp. ETL-1982 are shown in figures 4a-4d. Additional glucose concentration enhanced the decolorization of Reactive Red by Bacillus spp. ETL-1982 (Figure 4a). However, lack of glucose inhibited the decolorizing activity of Bacillus spp. ETL-1982 since only 26.72% color removal was observed after 120 h incubation. In experiments with glucose supplementation, Bacillus spp. ETL-1982 exhibited strong decolorizing activity with about 90% decolorization extent in 48 h, except that when the glucose concentration was 0.5 g l-1 or 12 g l-1, the decolorization efficiencies (64.19% and 67.23% in 120 h, respectively) were much lower. The reason why low decolorization extent appeared when the glucose concentration was 0.5 g l-1 may be that low glucose concentration could not meet the growth requirements of the bacteria. When the glucose concentration was much higher, such as 12 g l-1, the bacteria could utilize glucose preferentially, thus resulting in lower decolorization extent. The optimum pH for Bacillus spp. ETL-1982 for dye decolorization was pH 9.0. Bacillus spp. ETL-1982 was also capable of decolorizing the dyes at a wider pH range of 6-11 with an appreciable efficiency on either side of the pH optima. Majority of the azo dye reducing species of Bacillus and Pseudomonas reported [25,27,28] so far were able to reduce the dye at pH near neutrality However, optimum pH for the Bacillus spp. ETL-1982 mediated dye degradation was found to be 9.0 (Figure 4b), this observation is of extreme significance in deploying Bacillus spp. ETL-1982 for bioremediation. The decolorization of the dye Reactive Red was studied within a temperature range of 20 to 45°C. The optimum temperature for the growth of Bacillus spp. ETL-1982 for dye decolorization was 37°C. The dye decolorization efficiency of Bacillus spp. ETL-1982 decreased on either side of temperature optima (Figure 4c). Process With increase of the initial dye concentration, the decolorization extent over the same time interval decreased. When the effect of different initial dye concentrations of Reactive Red on decolorization was observed using 25, 50, 100, 200 and 500 mg l-1, the required times to reach a maximum decolorization extent were 24, 24, 36, 36 and 108 h, respectively (Figure 4d). It was reported that dye decolorization can be strongly inhibited when a high concentration dyestuff was used to examine the poisonous effect of the dye on the degrading microorganisms [27]. To accurately appraise the decolorizing ability of Bacillus spp. ETL-1982 decolorization was investigated by assessing the amount of dyes decolorized. Although the decolorization extent was only 41.69% at 1000 mg l-1 dye after 120 h incubation, 416.9 mg dye had been degraded, only 42 mg less than the decolorization at 500 mg l-1. This result indicated that Bacillus spp. ETL-1982 showed high decolorizing performance even in high initial dyestuff concentrations. |
| Decolorization manner of Bacillus spp. ETL-1982 against reactive red |
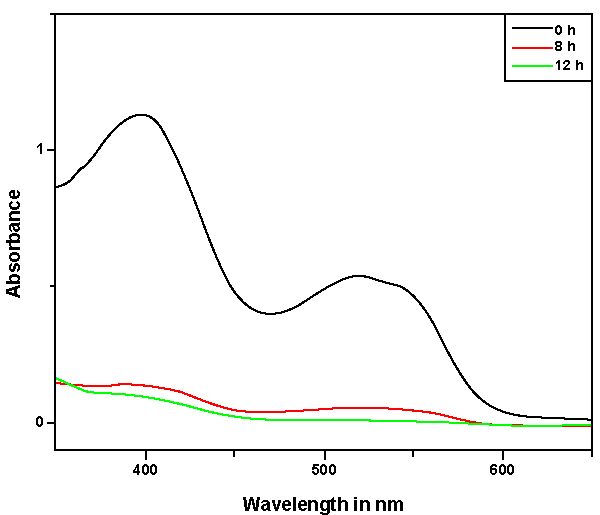
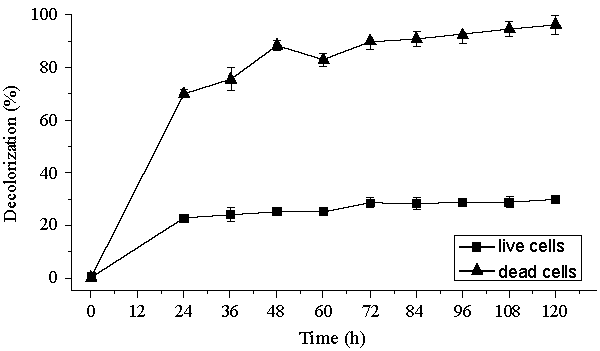
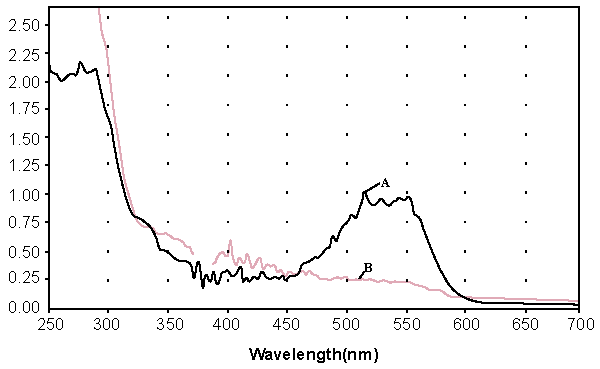
| Decolorization of dyes may take place by adsorption [29] or degradation [30]. In the case of adsorption, dyes are only adsorbed onto the surface of bacterial cells, whereas new compounds come into being when dyes are degraded by bacterial enzymes during the degradation process. In adsorption, examination of the absorption spectrum will reveal that all peaks decrease approximately in proportion to each other. If the dye removal is attributed to biodegradation, either the major visible light absorbance peak will completely disappear or a new peak will appear [31]. Dye adsorption can be also easily judged by an evidently colored cell pellet, whereas those retaining their original colors are accompanied by the occurrence of biodegradation [13]. In the cultures with addition of heat-killed bacterial cells, only 25% decolorization extent was recorded after 96 h incubation, which might be due to the adsorption by dead bacterial cells, and this was also confirmed by the presence of colored cell pellets. In the control culture, 96% decolorization extent was achieved in 96 h (Figure 5) and the cell pellets were not pigmented. Meanwhile, a UV–vis spectral scan (250–700 nm) of supernatants after decolorization showed that the maximum absorbance wavelength in visible spectra shifted from 560 nm to 400 nm in control cultures (Figure 6). The great changes occurring both in UV and visible spectra indicated that the molecular structure of Reactive Red changed evidently after decolorization. The red color of Reactive Red was caused by the conjugated structure of azo bonds (chromophore) and amino group. It could be presumed that the azo bonds cleaved during the reaction which indicated that the primary chromophore was destroyed. The absorbance peak at UV spectra did not disappear in the end of decolorization, which indicated that Reactive Red was not completely mineralized while some new metabolites formed in the culture. These results provided obvious evidence of biodegradation of reactive dyes by Bacillus spp. ETL-1982 in the decolorization process, and also supported the earlier conclusion that decolorization by bacteria is due to biodegradation, rather than inactive surface adsorption [12,32-34] |
| Conclusions |
| In this study, a decolorizing bacterial strain, Bacillus spp. ETL- 1982, was isolated from activated sludge. Bacillus spp. ETL-1982 showed decolorizing activity through a degradation mechanism rather than adsorption, and it could tolerate high concentrations (up to 1000 mg l-1) of Reactive Red. With high degradative and decolorizing activity against various azo dyes commonly used in the textile industries, it is proposed that Bacillus spp. ETL-1982 has a remarkable practical application potential in the biotransformation of various dye effluents. |
| Acknowledgements |
| Authors are highly thankful to the management of Enviro Technology Limited., Ankleshwar, Gujarat, India for allowing us to carry our such a noble work for the sustainable environment. |
References
|
Figures at a glance
 |
 |
 |
 |
 |
| Figure 1 | Figure 1a | Figure 1b | Figure 2 | Figure 3 |
 |
 |
 |
 |
|
| Figure 4a | Figure 4b | Figure 4c | Figure 4d | |
 |
 |
|||
| Figure 5 | Figure 6 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 13597
- [From(publication date):
March-2013 - Nov 23, 2024] - Breakdown by view type
- HTML page views : 9123
- PDF downloads : 4474
