Research Article Open Access
Biodegradation and Decolorization of Acid Red by Acinetobacter radioresistens
| Mohandass Ramya *, Iyappan S, Manju A and Jiffe John S | |
| Department of Genetic Engineering, School of Bioengineering, SRM University, Kattankulathur 603203, India | |
| Corresponding Author : | Ramya Mohandass Department of Genetic engineering School of Bioengineering SRM University Kattankulathur 603203, India E-mail: ramya.mohandass@gmail.com |
| Received August 12, 2010; Accepted September 28, 2010; Published October 02, 2010 | |
| Citation: Ramya M, Iyappan S,ManjuA, Jiffe JS (2010) Biodegradation and Decolorization of Acid Red by Acinetobacter radioresistens. J Bioremed Biodegrad 1:105. doi: 10.4172/2155-6199.1000105 | |
| Copyright: © 2010 Ramya M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Acid red decolorizing bacteria was isolated and identified as Acinetobacter radioresistens.The effect of operation parameters such as medium composition, pH, temperature, dye concentration on the decolorization of acid red was studied and the products of degradation were analyzed and confirmed using LC–MS analysis. The reductive cleavage of azo bond was catalyzed by azoreductase, the key enzyme for the azo dye degradation. The catalytic reduction of acid red 37 by purified azoreductase in the presence of NADH as electron donor was studied and the products of degradation were determined as 1-{3-amino-5-[(aminoxy)sulfonyl]phenyl}ethanol and 7,8-diamino-3[(aminoxy)sulfonyl]naphthalene-1-ol.
| Keywords |
| Decolourization; Degradation; Azo Dyes; Azoreductase; LC-MS |
| Introduction |
| Large amounts of dyes are annually produced and applied in many different industries, including textiles, cosmetics, paper, leather, pharmaceutical and food [1]. The three most common groups are azo, anthraquinone and phthalocyanine dyes. Most of these dyes are toxic and carcinogenic [2]. Disposal of these dyes into the environment causes serious damage, since they may significantly affect the photosynthetic activity of hydrophytes by reducing light penetration and also they may be toxic to some aquatic organisms due to their breakdown products [3]. The degradation of azo dyes produces aromatic amines, which are carcinogenic, and mutagenic [4]. Recently, several reports appeared showing that the microorganism has ability, not only to decolorize dyes but also detoxify it [5,6]. |
| Microbial degradation and decolorization is an environment friendly and cost-competitive alternative to chemical decomposition processes. Many microorganisms belonging to different taxonomic groups of bacteria, fungi, actinomycetes and algae have been reported for their ability to decolorize azo dyes [7]. Acid red is one of the azo dye which has large consumption rate in textile industry. |
| Acinetobacter sp reported to degrade various dyes in the earlier reports [8,9]. A. calcoaceticus a textile soil isolate reported to degrade diverse dyes present in the textile effluent, into nontoxic metabolites [10]. Many researchers reported that Acinetobacter radioresistens has efficiency to degrade various aromatic hydrocarbons [11,12].However no report on dye degradation efficiency and enzyme characteristics of this organism Acinetobacter radioresistens. This is the first report on the efficiency Acinetobacter radioresistens in degradation of dye. Various parameters such as agitation, temperature, pH and different dye concentration required to achieve maximum dye decolorization were standardized. The organism effectively degrades the dye by the partially purified enzyme azoreductase. The enzyme and whole cell culture tested for their efficiency to degrade acid red 37 and the intermediates products formed were analyzed by liquid chromatography and mass spectroscopy (LC–MS). |
| Materials and Methods |
| Dyes and chemicals |
| Acid red 37 (Catologue number- 210633, Ashwin colour company Pvt. Ltd, Chennai, TamilNadu, India) was used to prepare the stock solution, which was filter sterilized and added to the growth medium at required concentrations. All the chemicals used were of analytical grade and procured from Himedia Pvt. Limited, Mumbai. The materials required for PCR amplification (Taq polymerase, dNTPs, primers) were obtained from Bioserve Biotechnologies India Pvt. Ltd, Hyderabad, India. |
| Isolation and screening of dye decolourising bacterium |
| Soil samples contaminated with textile dye effluent were collected from different vicinities in Chennai and were used as a source to isolate morphologically distinct bacterial colonies capable for decolourising acid red 37. The soil samples were subjected to enrichment culture technique using nutrient broth amended with increasing concentrations of dye (100 mg/L 500mg/L) with incubation time of 24 h at 37°C. Repeated transfers were carried out to isolate stable dye decolourising strains and the isolated strains were stored at 15°C for further use. The high decolourising bacteria were screened by performing a decolourisation assay with acid red using UV- VIS spectrophotometer (Ultrospec 2100, Amersham Biosciences, UK) at its respective λmax 502 nm. |
| The percentage decolourisation was calculated using the following formula- |
 |
| Initial absorbance - Absorption at 0 h of incubation. |
| Final absorbance - Absorption at the time of incubation. |
| Molecular identification of the isolate |
| Genomic DNA from the isolated organism was isolated and its presence was checked by running in agarose gel (0.8%) stained with ethidium bromide. Amplification of 16S rDNA sequence by polymerase chain reaction was done using thermal cycler (Gene Amp® 2720). 16S r DNA universal primers used for PCR reaction [13]. The reaction mixture of total volume of 30µL consisted of 3µL of 10X Buffer, 1µL of 10mM dNTPs, 1µL of 16S rDNA primer (5 picomole/µL), 3U/µL of Taq Polymerase, 5µL of template DNA (280 ng/ml) and 19µL of sterile distilled water. The PCR reaction was set to initial denaturation of 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for one minute, annealing at 55°C for one minute, extension at 72°C for one minute and final extension at 72°C for 10 minutes [14]. The amplified products were stained with 0.5 µg/ml ethidium bromide and loaded on 0.8% agarose gel, and the DNA fragments were separated at 100V and documented. The amplified product was subjected to cycle sequencing using ABI 3130 XL (Genetic Analyser, Applied Biosystems, USA). The sequence was deposited to Genbank (http://www.ncbi.nlm. nih.gov/Genbank). The nucleotide sequence analysis of the sequence was performed at BlastN site at NCBI server (http://www.ncbi.nlm. nih.gov/BLAST). The alignment of the sequences was performed by using Clustalw program V1.82 at European bioinformatics site (http:// www.ebi.ac. uk/clustalw). The Phylogenetic tree was constructed by the neighbour joining method using Kimura-2-parameter distances in mega 2.1 software [15]. |
| Effect of physico-chemical factors on decolourisation |
| Mid log phase culture of the isolate was inoculated into LB media supplemented with acid red. The effect of various physicochemical factors (dye concentration, temperature, pH, oxygen) on the decolorization of acid red by the isolate was studied in detail. Decolorization of acid red by the isolate was carried out at various dye concentrations (50, 100, 200, 400 mg/L), temperature (20°C, 40°C, 60°C, 80°C) and pH (2-9). Decolorization experiment was also carried out under aerobic and anaerobic conditions. All decolorization experiments were performed in triplicates. Abiotic controls with bacteria were always included. |
| Characterization and purification of the intracellular azoreductase enzyme |
| Cells from the mid log phase culture were harvested by centrifugation at 10000 rpm for 10 minutes at 4°C. Pellets were disrupted by sonication at 40% power for 6 minutes. The cell lysate was subjected to fractionated ammonium sulfate precipitation at 40% saturation to remove impurities, followed by 70% saturation in a second step to precipitate the azoreductase. After 24 h, the precipitated protein is centrifuged for 10 minutes at 10000 rpm at 4°C and the pellet was dissolved in equal volume of 50mM potassium phosphate buffer (pH 7.2). Ammonium sulfate precipitated sample was then desalted by dialysis against phosphate buffer (50mM, pH 7) overnight under room temperature. 2mL of the resulting solution was fractionated by anion exchange chromatography using DEAE sephadex column installed in an Amersham Pharmacia Biotech AKTA purifier; pump P-900, monitor pH/C- 900, monitor UV-900, auto sampler Frac-950. Elution buffer (sodium phosphate buffer containing 1M NaCl was set to a gradient of 100% for 150 minutes. Proteins were eluted at a flow rate of 1.5mL/minute [16]. The fractionated sample was concentrated using protein purification column. Active fractions were collected and stored as the purified enzyme preparation. |
| Assay for azoreductase activity |
| Assays were carried out in cuvettes with a total volume of 1mL using Ultrospec 2100 UV-VIS Spectrophotometer (Amersham Biosciences). The reaction mixture consists of 400µL of potassium phosphate buffer with 200µL of sample and 200µL of acid red 37 (100mg/L). The reaction was started by addition of 200µL of NADH (7mg/mL) and was monitored photometrically at 502 nm. The linear decrease of absorption was used to calculate the azoreductase activity [17]. One unit of azoreductase can be defined as the amount of enzyme required to decolorize 1µmol of acid red per minute. The effect of pH on azoreductase activity was determined by incubating the reaction mixture at pH values ranging from 4 to 9. The optimum temperature for enzyme activity was determined by conducting the assay at various temperatures from 20°C to 80°C in 50mM potassium phosphate buffer (pH 7).The relative activity of azoreductase at each temperature was determined. |
| Liquid chromatography and mass spectroscopy analysis (LCMS analysis) |
| About 100ml of acid red (200mg/L) containing LB Media treated with the isolate and the purified enzymes was extracted with equal volume of ethyl acetate at various time intervals (0, 24, 48 h).The extract was evaporated in a vacuum evaporator (Buchii R 124, Germany) and used for LC- MS analysis. The powdery residue was then dissolved in acetonitrile (HPLC Grade). LC-MS analysis was performed using Finnigan model Mass Spectrometer (Thermo Electron Corporation, USA) using C-18 column from Waters. The cartridges were conditioned with pure acetonitrile, washed with deionized water (0.1% Formic acid) and the elution took place with 70% acetonitrile, containing 0.1% formic acid. The flow rate was 0.8 ml/ min. The ion trap detector with atmospheric pressure electro-spray ionization (API-ESI) source was used for quantification in negative ionization mode. Operating conditions were dry with temperature of 325°C, Capillary voltage 3500V, Nebulizer 14 psi, dry gas Helium 5.0 l/min. Ion trap full scan analyses were conducted from m/z 200- 1400 with an upper full time of 300 minutes. The nebulizer gas flow and the curtain gas flow (Nitrogen gas) were set at 10 and 8 psi. The ion spray, orifice and ring voltage were set at +4800, 40, +70 V respectively. Instrumentation control of data acquisitions were performed with data analysis MS (X caliber, USA). |
| Results and Discussion |
| Screening and Isolation of dye decolorizing bacteria from soil |
| From the serially diluted soil sample, 16 morphologically distinct bacterial colonies with decolourizing zones were isolated. All the 16 bacterial isolates were tested for their dye decolourising efficiency by liquid decolorization assay. Using 500 mg/L of acid red 37, five Bacterial isolates - R6, R7, C5, C7 and C8 showed decolorization percentage of more than 70% at 48 h of incubation. The highest decolorization percentage was showed by C8 (87.2 ± 0.45). This bacterial isolate was selected and used for further studies. |
| Molecular characterization and identification of the isolate |
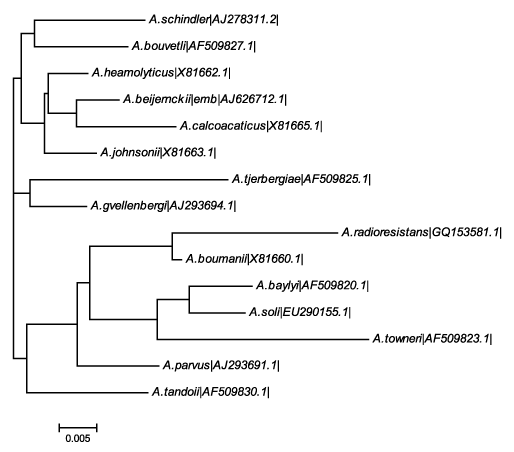
| DNA was isolated from the screened bacterial isolate C8. 16S r DNA sequence of the colony C8 was amplified using universal primers. The size of the amplified band was determined to be approximately 1.5 Kb. Each of the amplified DNA was further purified by DNA Elution kit (Bioserve Biotechnologies India Private Limited). The purified bands were subjected to cycle sequencing and BLAST to determine the organism. The isolate C8 was identified as Acinetobacter radioresistens by BLAST analysis (www.ncbi.nlm.nih. gov/BLAST/) in the nucleotide sequences database. The sequence was submitted to genbank with the accession number (GQ 153581.1).The phylogenetic tree shown in Figure 1 clearly shows that the isolated strain was closely related to A. boumanii. With in this family it shows distinct from the A. schindler. Phylogenetic analysis using other methods of tree building also supported this grouping with high bootstrap values. |
| Effect of physico-chemical factors on decolourisation |
| Maximum decolorization of 90% was observed in pH from 6 to 7 (table 1). Rate of decolorization decreased at lower pH (4-5) and at higher pH (8-9). No decolorization was obtained at lesser pH 1-2. K. pneumoniae RS-13 was found to completely degrade Methyl Red at pH 6.0-8.0 while Acetobacter liquefaciens S-1 completely degraded Methyl Red at pH 6.5 [18]. E. coli and P. luteola both exhibited best decolorization rate at pH 7.0 with constant decolorization rates up to pH 9.5 [19]. The isolate showed higher decolorization percentage when the medium was incubated at 40-50°C. The isolate is capable of decolorizing the dye up to 400mg/L. Lee et al. [20] stated that the increase in the concentration of the dye, the ability of the organism to decolorize decreased. In contrast to this decolorization was not affected by increasing the dye concentration (50mg-400mg/L). As indicated in previous studies [21], the chemical structure characteristics (e.g., resonance and inductive effects) and reactivity of dyes strongly determined color removal efficiency of bacterial decolorizers. |
| Characterization and purification of azoreductase |
| The enzyme was assayed for extracellular activity. As there was no significant activity observed, intracellular release of the enzyme was performed. After each step of purification the activity was assayed and it was found that the specific activity of the enzyme increased after each step of purification. Enzyme activity in the crude cell extract was found to be 0.0009U/mg. After ammonium sulfate precipitation and FPLC, the activity increased to 88.49U/mg. Table 2 gives the activity of azoreductase after each step of purification. The optimum pH found for the activity of azoreductase enzyme was pH 7 with an activity of 0.0003U/mg. The optimum temperature was found to be 40°C with an activity of 0.00072U/mg. |
| Biodegradation of Acid Red 37 by LC-MS |
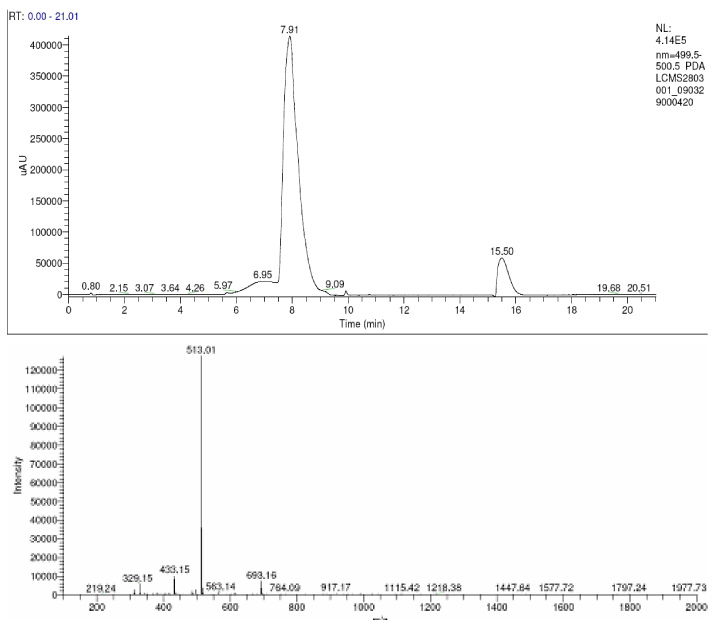
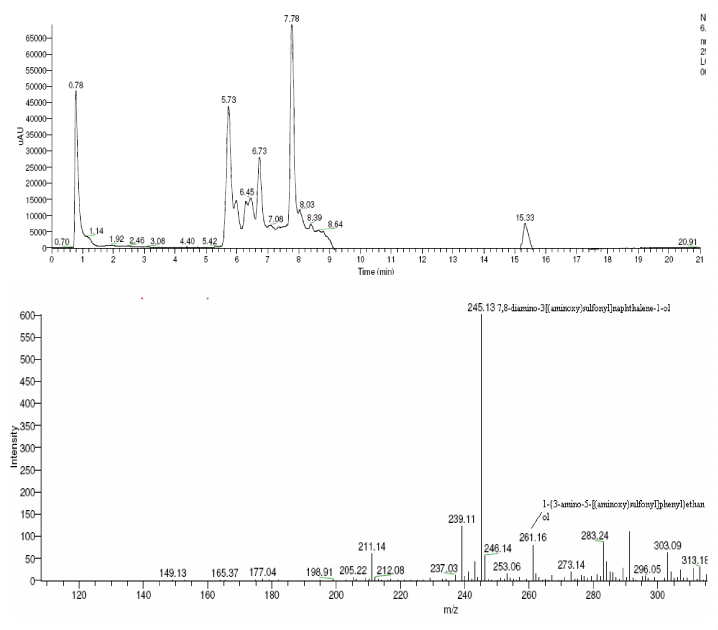
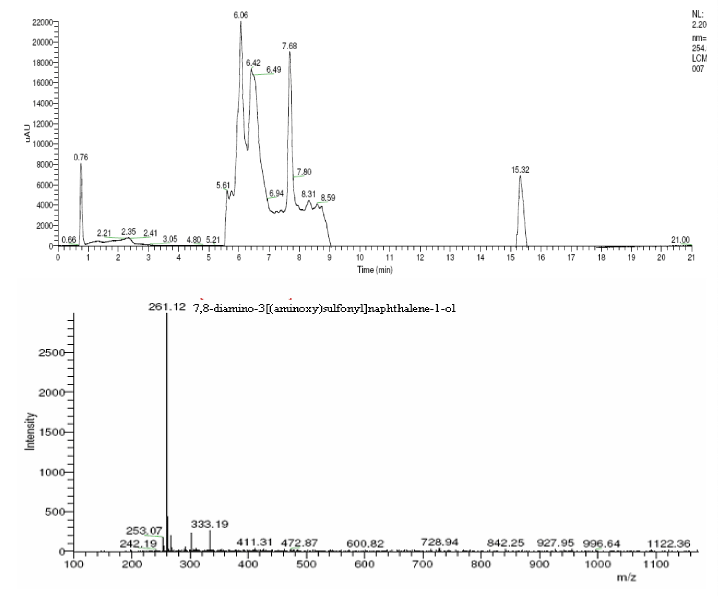
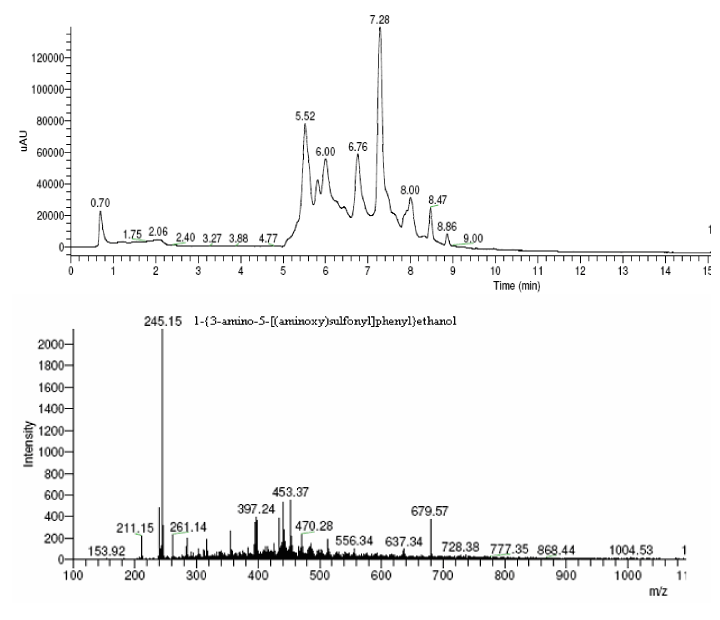
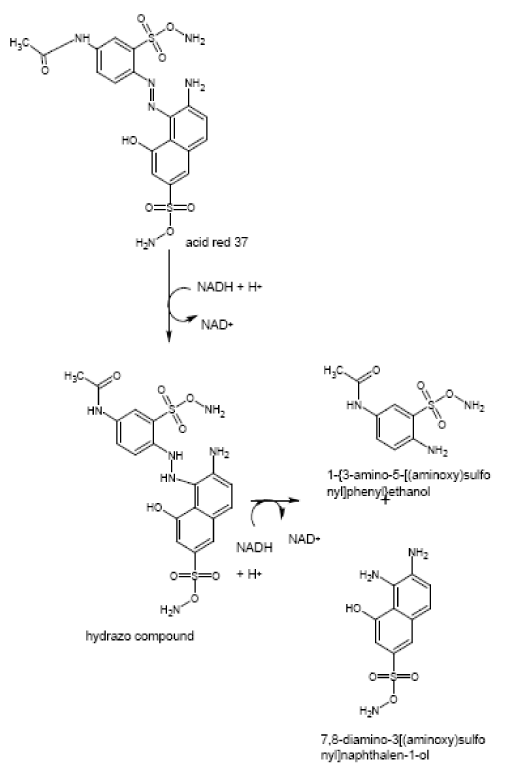
| The products of degradation were analyzed and the spectrum of LC-MS of the 0h, 24 h sample and 48 h sample is shown in the Figure 2. The chromatogram of acid red at 0th hour showed a peak at retention time of 8.01. The mass spectrometric peak at this retention time showed m/z ratio of 513.01 (Figure 2a). In the case of decolourized culture(24h) of acid red, the mass spectrometric peak at retention time of 5.71, showed a significant m/z ratios of 261(24h),which corresponds to 1-{3-amino-5-[(aminoxy)sulfonyl] phenyl}ethanol (Figure 2b). A new peak with the retention time of 5.9, m/z ratios of 245.15 was formed at 48h which corresponds to 7,8-diamino-3[(aminoxy)sulfonyl]naphthalene-1-ol (data not shown). In case of decolorization with the enzyme (Figure 2c) the mass spectrometric peak at retention time of 6.6(m/z ratio 245.16) was seen at 3 h of incubation with the enzyme. Figure 2d shows that at 5h of incubation new peak with the retention time of 5.8(m/z ratio of 261.11) was formed. These mass values corresponds to 1-{3-amino- 5-[(aminoxy) sulfonyl] phenyl}ethanol and 7,8-diamino-3[(aminoxy) sulfonyl]naphthalene-1-ol . In the degradation pathway, hydrogen ions are donated to the azo group of acid red 37 and a hydrazo compound is formed which further reduced to aromatic amines. As shown in Figure 3, the possible mechanism of degradation could be cleavage of azo bond resulting in the formation of degraded products like 7, 8-diamino- 3[(aminoxy)sulfonyl]naphthalene-1-ol and 1-{3-amino-5-[(aminoxy)sulfonyl]phenyl}ethanol through two step NADH dependant reduction mechanism. The complete catabolic pathway of degraded products has to be studied. However, these amino derivatives/intermediates are reported to be degraded/ transformed under aerobic conditions [22]. |
| Conclusion |
| The results obtained in this paper were very promising since azoreductase from Acinetobacter radioresistens could able to degrade the acid red 37. The partially purified enzyme azoreductase was found to degrade the dye via symmetric cleavage of its azo bond. The culture exhibited good decolorization ability at pH from 6 to 8 and temperature from 30 to 50°C. Decolorization efficiency was not affected by the concentration of the dye. However, more studies in order to study the heterologous expression of this enzyme are underway in our laboratory. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2a | Figure 2b |
 |
 |
 |
| Figure 2c | Figure 2d | Figure 3 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 16349
- [From(publication date):
October-2010 - Jan 09, 2026] - Breakdown by view type
- HTML page views : 11465
- PDF downloads : 4884
