Research Article Open Access
Enrichment of Phenol Degrading Moderately Halophilic Bacterial Consortium from Saline Environment
| Krishnaswamy Veena Gayathri* and Namasivayam Vasudevan | |
| Centre for Environmental Studies, Anna University, Chennai-6000 25, India | |
| Corresponding Author : | Krishnaswamy Veena Gayathri Centre for Environmental Studies Anna University, Chennai-6000 25, India Tel: 0-91-044-222359029 E-mail: veenagayathri@yahoo.com |
| Received: July 24, 2010; Accepted: September 28, 2010; Published: October 01, 2010 | |
| Citation: Gayathri KV, Vasudevan N (2010) Enrichment of Phenol Degrading Moderately Halophilic Bacterial Consortium from Saline Environment. J Bioremed Biodegrad 1:104. doi:10.4172/2155-6199.1000104 | |
| Copyright: © 2010 Gayathri KV, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
A versatile, moderately halophilic bacterial consortium was developed for the biodegradation of phenolic compounds under saline conditions. The bacterial consortium was isolated from mixtures of soil from phenol contaminated sites and as well as from areas having proximity to saline environment. The isolated moderately halophilic bacterial consortium utilized different phenolic compounds as sole source of carbon source. Phenol was utilized by the consortium at a range of salt concentrations from 10- 150 g/L NaCl where the optimum degradation was achieved at 50 g/L of NaCl. The bacterial consortium utilized up to 300 mg/L of phenol most effi ciently than the individual strains present in the consortium under saline conditions. The 16S r-RNA gene analysis and biochemical tests showed that the bacterial consortium contained six bacterial strains, which were identifi ed as Bacillus cereus, Arthrobacter sp., Bacillus licheniformis, Halomonas salina, Bacillus pumilus and Pseudomonas aeruginosa. Such moderately halophilic bacterial consortium might be useful for the treatment of industrial saline wastewater, particularly, in environments contaminated with phenolic wastes.
| Introduction |
| Phenol is one among the most prevalent forms of chemical pollutants, because it is toxic even at low concentrations, and also its presence in natural waters can lead further to the formation of substituted compounds during disinfection and oxidation processes. Phenol is a toxic industrial compound whose presence in the environment poses significant risks to aquatic biota to fish at relatively low concentrations of (5-25 mg/L) [1]. Phenols are major pollutants of industrial wastewaters since they are commonly used in many industries such as oil refining, coke conversion, pharmaceutical and resin manufacturing plants. |
| Hypersaline wastewaters are defined as brines which contain organic compounds and at least 3.5% w/v total dissolved solids (TDS). These waters are generated during the manufacture of chemicals such as pesticides, herbicides, polyhydric compounds, organic peroxides and pharmaceuticals [2]. Saline and hyper-saline environments are frequently contaminated with organic compounds as a result of industrial activities [3,4]. Contamination of these habitats constitutes a serious environmental problem mainly due to the high toxicity exhibited by aromatic hydrocarbons. |
| In most cases, biodegradation constitutes the primary mechanism for the organic contaminant removal. However, biodegradation processes are difficult to perform under saline conditions [3-5]. In addition, it is known that the traditional pollutant biodegradation is less efficient or does not function when salinity increases above that of the sea [4]. Hence, it is a challenging issue in the treatment of wastewater in petrochemical, pesticide, pharmaceutical, mining and other industries for effective biological treatment. One alternative is the use of halophilic organisms which are adapted to live in such saline conditions. |
| The complex organic and inorganic composition of many industrial waste brines can make them difficult to treat. High salinities or wide salinity gradients may limit microbial degradation of organic compounds and make conventional biodegradation techniques ineffective. In these cases halotolerant or halophilic microorganisms must be used as they have the capability to grow in salinity. Moderately halophilic bacteria are extremophilic microorganisms adapted to live in saline environments. These halophiles grow optimally, in media containing between 3% and 15% NaCl [6], although the main characteristic of this group of organisms is their ability to grow in a very wide range of salt concentrations. They constitute a very interesting group of microorganisms with a great potential for use in biotechnology. |
| Phenols are major pollutants of industrial wastewaters since they are commonly used in many industries such as oil refining, coke conversion, pharmaceutical and resin manufacturing plants. The degradation of phenol by both pure and mixed cultures under nonsaline conditions, however, very few reports have been documented, especially by moderately halophilic bacteria on biodegradation of phenol [7-10]. Salt tolerant phenol degrading organisms isolated from the Amazonian soil was reported [11]. The degradation potential of a halophilic bacterium Halomonas organivorans was studied to catabolise different concentrations of low molecular weight aromatic compounds at 10% NaCl concentration [12]. Wang et al [13] indicated that there was almost no effect on the growth of Arthrobacter sp. and the degradation of mixtures of phenol (200 mg/L) and p-cresol (100 mg/L) with less than 5 % NaCl in 88 h. The effects of NaCl on biodegradation of phenol have been well documented; the use of consortium in degradation phenolic compounds in saline industrial effluents has been ignored. In the present study, we have enriched and characterized a phenol degrading halophilic bacterial consortium from saline environment to degrade phenolic compounds at wide range of salt concentrations. |
| Materials and Methods |
| Enrichment and isolation of the bacterial consortium |
| Soil samples were collected from different ecosystems in Chennai such as salt pan, Puliket marine back water lake; Sea harbour (Chennai), phenol containing wastewater, tannery affected soils and soil from sea food industries. The bacterial consortium were enriched in mineral salts medium of NaCl 10.0-100.0, KH2PO4 0.25, NH4Cl 1.0, Na2BO7 2.0, FeCl3 0.0125, CaCl2 0.06 and MgCl2 0.05 all the salts as gram in one litre. The medium was supplemented with a specified amount of added NaCl, and 10 mg of yeast extract adjusted to pH -7. [14]. The medium was autoclaved, cooled to room temperature and was amended with phenol (50 mg/L) through a sterile filter (0.45 µm) in 250ml Erlenmeyer flasks. The bacterial consortium was enriched on phenol (50 mg/L) in mineral salts medium with 30 g/L NaCl concentration. The culture was then transferred, four times after every four days. Increase in cell count of the bacterial consortium from 105 to106 cfu/mL with phenol was taken as a confirmation for utilization of phenol. Once the consortium was fully acclimatized to phenol, its ability was analysed to grow on other phenolic compounds (Catechol, o-Cresol, m-Cresol, p-Cresol) and on substituted chlorophenols (2-Chlorophenol (2-CP), 4-Chlorophenol (4-CP), 2,4-Dichlorophenol (2,4-DCP), 2,4,6-Trichlorophenol (2,4,6-TCP) and Pentachlorophenol (PCP)). The NaCl concentrations used in the mineral salts medium were 1%, 3%, 5%, 7%, 10% and 15%. The chemicals and reagents used in the study were analytical grade. |
| Preliminary studies on phenolic compounds degradation |
| The bacterial consortium isolated from the saline environment was grown and the plate count (cfu/mL) was checked daily. Cell morphology and the motility of cells in exponentially-growing liquid cultures were examined on freshly-prepared wet mounts by light microscopy. Plate counting (cfu/mL) was done on nutrient agar medium. The consortium was studied for its growth on different phenolic compounds as the sole carbon source. For the degradation study, mineral medium containing phenolic compounds was inoculated with the bacterial consortium/ bacterial strain. Different conditions used for the degradation of phenol were (i) medium + phenolic compound + bacterial consortium; (ii) medium + phenolic compound and (iii) medium + bacterial consortium, with (ii) and (iii) serving as controls. The bacterial consortium was added to the medium at concentrations of 105–106 cfu/mL. The culture, in duplicate, was incubated at 37°C with shaking at 150 rpm and extracted every 24 h for 5 days. Each culture was extracted twice with ethyl acetate (v/v) after acidification to pH 2.5 with 1N HCl. The extracts were filtered through anhydrous sodium sulphate and condensed to 1 mL with a rotavapour (Buchi, Germany) for further gas chromatographic analysis. |
| Total protein analysis |
| For analysis of total cell protein, samples were centrifuged at 12,000 rpm for 10 minutes and washed with fresh (substrate-free) mineral medium, then centrifuged and washed few times to remove the substrate. The pellet from each sample was then disrupted by sonication at 30% amplitude for a total of 3 minutes (1.5min x 2) in an ice-water bath. To 0.5 mL of sample was added to 0.5 mL Coomassie Blue protein dye and the absorbance at 595nm were measured. Total protein concentration was determined by calibration with bovine serum albumin standards according to Bradford. |
| Gas chromatographic analysis |
| The ability of the consortium to utilize phenol as sole carbon source was determined by growing it in the mineral medium containing different concentrations of phenol from 50 mg/L to 250 mg/L. The cell suspensions were clarified by centrifugation at (10,000rpm for 15 min, 6°C). The culture supernatant was extracted with dichloromethane and filtered through a 0.2µm Gelman filter acro disc, prior to analysis in gas chromatograph (Chemito GC Model No 1000) equipped with FID detector and capillary column (Varian Chromopak capillary column CP SIL 8CB, 30m X 0.32mm with detection limit of 10 ppt of the compound. Nitrogen was used as a carrier gas, injector temperature was 220°C, detector temperature was 250°C and the oven temperature of the column was maintained at 150°C. |
| Identification of phenol degrading bacterial strains |
| The individual bacterial strains were separated from the consortium, which was used in the degradation of the phenolic compounds. The bacterial strains present in the consortium were examined using conventional biochemical tests initially. Molecular identification of the bacterial strains was done by 16S rRNA sequencing. |
| Scanning electron microscopy |
| The sample preparation for SEM was carried out according to the method of Prior and Perkins [15]. The isolates were grown on mineral salts medium (MSM) for 24h, centrifuged and pellets were immediately resuspended in 2% glutaraldehyde with 0.05M phosphate buffer and 4% sucrose (pH 7.3). Pellets were placed on aluminium foil disks, air dried, platinum coated and examined under SEM (JEOL JSM- 6360). |
| Biochemical characterisation |
| The bacterial strains present in the consortium were isolated and grown separately. Initially, Gram staining and motility test were performed after which biochemical characterization was done with KB003: Hi24 “Enterobacteriaceae Identification Kit”, (Himedia, India) to identify the phenotypic characters of the bacterial strains. The identification strip consists of 24 slots with different tests (Oxidase, ONPG, Lysine decarboxylase, Ornithine, Urease, Phenylalanine deamination, Nitrate reduction, H2S Production, Citrate Utilization, Methyl red, Voges Proskaures, Indole production, and growth on Malonate, Esculin, Arabinose, Xylose, Adonitol, Rhamnose, Cellobiose, Melibiose, Saccharose, Raffinose, Trehalose, Glucose and Lactose) to which 10µL of 24h old culture was added. After 24 h incubation at 37oC, the colour change observed was accounted for positive/negative result. Genus level identification of the unknown bacterial strains were accomplished by the using Bergey’s Manual of Systematic Bacteriology (2005) to ascertain the existence of variable biochemical test results for each strain. |
| 16S rRNA partial gene sequencing |
| Chromosomal DNA was isolated from the pure strains of the consortium by the standard phenol/chloroform extraction method [16]. The 1.5 kilo base partial sequence of the 16S rRNA gene was amplified from the chromosomal DNA using polymerase chain reaction (PCR) with universal Eubacteria- specific primers 16F27 (5’- CCA GAG TTT GAT CMT GGC TCA G-3’) and 16R1525XP (5’-TTC TGC AGT CTA GAA GGA GGT GWT CCA GCC-3’) [17]. The PCR conditions used were an initial denaturation at 94°C for two minutes, followed by 35 cycles of denaturation at 95°C for one minute, annealing at 55°C for one minute, and extension at 72°C for one minute, and a final extension at 72°C for 10 minutes and sequenced on an ABI310 automated DNA sequencer using the Big Dye terminator kit (Applied Biosystems, Inc., Foster City, CA) [18]. The amplified 16S rRNA gene PCR products from these isolates were directly sequenced after purification by precipitation with polyethylene glycol and NaCl. The primers used to obtain the complete sequence of 16S rRNA gene of the isolates were the same as for PCR amplification (16F27N and 16R1525XP) [17]. |
| Sequence analysis |
| The sequences from forward or reverse amplification were aligned with16S rRNA sequence data set retrieved from the Ribosomal Database Project (RDP II; Michigan State University, East Lansing, MI) and the National Center for Biotechnology Information (Bethesda, MD) (http//:www.ncbi.nlm.nih.gov/BLAST). The similarity matrix was prepared using the similarity matrix calculator at the RDP II site. The presence of chimeric sequences was checked with the RDP Chimera Check program and by comparing independently the alignments at the beginning of each sequence, end of each sequence and the alignments of the entire sequence. The alignment of all the 16S rRNA gene sequences was done using CLUSTAL W program at the European Bio Informatics server (European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, United Kingdom) (http//: www.ebi.ac.uk/clustalw) to determine similarity value and distance matrix. The evolutionary history was inferred using the Maximum Parsimony method [19]. The bootstrap consensus tree inferred from 500 replicates was taken to represent the evolutionary history of the taxa analyzed [14]. The (Maximum Parsimony) MP tree was obtained using the Close-Neighbour-Interchange algorithm [20]. The boot strap consensus tree was constructed using 500 replicate aligned sequences by using maximum parsimony method and the phylogenetic analysis was conducted in MEGA4 software [21]. |
| Results and Discussion |
| Isolation of bacterial consortium from saline environment |
| Soil samples were collected from different saline ecosystems to isolate phenol degrading bacterial strains which could degrade phenol under saline conditions. During the isolation period, several bacterial strains co-existed in the consortium which could degrade phenol (50 mg/L) under saline conditions. After successive transfers, during the enrichment period the bacterial consortium contained six strains, which survived and degraded phenol as the sole carbon source. This condition could be due to selection within the population for specific bacterial strains, which are more tolerant to phenol, probably due to changes in the regulatory system, achieved by substrate specificity or over expression of a pre-existing system for degradation of related substrates [22]. Another reason could be when the enrichment culture is exposed to higher concentrations of the pollutant; there is further selection of microorganisms that are highly tolerant to the compound. Acquisition of degradative abilities by selective enrichment has been seen in laboratory ecosystems for many organic compounds and heavy metals [23,2]. |
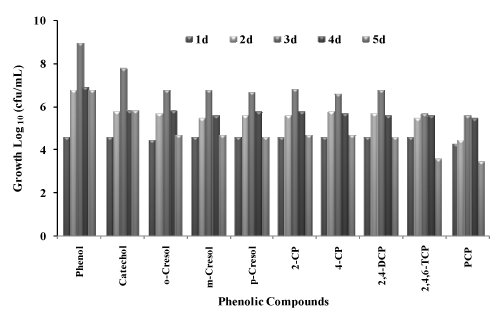
| Growth of bacterial consortium on different phenolic compounds |
| The bacterial consortium was studied for utilization of different phenolic compounds to be used as a sole source of carbon source and energy in the presence of 30 g/L NaCl concentration. Growth of the bacterial consortium on different substrate is given in table 1. As the consortium was isolated from phenol contaminated sites and acclimatized to grow on phenol during the enrichment period, the consortium was able to grow on all the selected phenolic compounds which are indicated by increase in cfu/ml on different phenolic compounds (Figure 1). The growth of the bacterial consortium was comparatively less on chlorinated compounds like 2,4,6-TCP and PCP. This may be due to the substituted chlorine atom attached to the substrate. The isolated bacterial consortium was able to grow on phenols (Phenol, Catechol, o-Cresol, m-Cresol, p-Cresol) and on substituted chlorophenols (2-CP, 4-CP, 2,4-DCP and 2,4,6-TCP) but the growth was very less on PCP. The growth of bacterial consortium on PCP was very slow, probably because of its recalcitrant structure with five chloride substitutes attached to the aromatic ring structure. The toxicity of phenolic compounds tends to increase with relative degree of chlorination [24]. This view was also supported by Saber and Crawford [25], where they reported that PCP was resistant to degradation because of its stable aromatic ring system and high chloride content. In present study, it was found that the growth of the consortium on PCP was inhibited with reduction in the viable cell count. This reason is in accord with Gonzalez [26] where they showed a decrease in the viability of the mixed culture with PCP under non-saline conditions. The isolated bacterial consortium was able to utilize phenol as the sole carbon source, were the cell count increased from 4 x 104 and reached the maximum of 9 x 108 at the end of 3rd day. Growth of the bacterial consortium on catechol showed that the viable cell count increased from 4 x 104 to 6 x 107 cfu/ml in 3 days. In the case of derivatives of phenol o-cresol, p-cresol, and m-cresol the viable cell count increased from 3 x 104 to 6 x 106, 4 x 104 to 6 x 106 and 4 x 104 to 5 x 106 cfu/ml respectively at the end of 3 days. When compared with phenols, chlorophenols gave less colony forming units, this may be due to the toxicity of chlorine atom present in the compounds. During the utilization of 2-CP and 4-CP the viable cell counts increased from 4 x 104 to 7 x 106 and 4 x 104 to 4 x 106 cfu/ml respectively. The cell count of the bacterial consortium with 2,4-DCP increased from 4 x 104 to 6 x 106 cfu/ml, the viable cell count for 2,4,6-TCP was comparatively less, where the count was 4 x 104 to 5 x 105 cfu/ml at the end of 3 days. The cell count was very less when the bacterial consortium was grown on PCP from 2 x 104 to 4 x 105 cfu/ml (Figure 1). This preliminary study on the growth of bacterial consortium on selected phenolic compounds showed that the consortium was able to utilize most of the phenolic compounds under aerobic conditions. Garcia et al [12] studied the degradation of low- molecular- weight aromatic compounds (benzoic acid, p-hydroxy benzoic acid, cinnamic acid, phenylacetic acid, p-coumaric acid, ferulic acid, salicylic acid) by a group of halophilic bacteria. When the isolates were enriched on phenol, they were able to utilize a greater number of aromatic compounds than the rest of the isolates enriched on other aromatic compounds other than phenol, showing their wider substrate specificity. But the mixed isolates were unable to utilize p-cresol. In present study the isolated bacterial consortium enriched on phenol was able to utilize most of the selected phenolic compounds including chlorophenols. |
| Identification of the consortium |
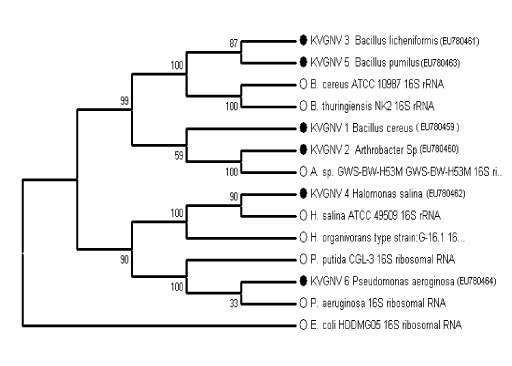
| Consortium consisted of 6 strains of bacteria of which 4 strains were Gram positive and two strains were Gram negative and all the six strains were motile. The biochemical and physiological characteristics of the organisms are given in table 1. Scanning electron microscopic photograph of the consortium is the given in the Figure 2. The identification of bacteria was carried out based on phenotypic characteristics, such as Gram staining, morphology, motility and nutritional requirement. Further analysis with the 16S r DNA gene sequence identified the 6 isolates as KVGNV1- Bacillus cereus, KVGNV2- Arthrobacter sp., KVGNV3- Bacillus licheniformis, KVGNV4- Halomonas salina, KVGNV5- Bacillus pumilus and KVGNV6- Pseudomonas aeruginosa. Figure 3 shows the phylogenetic relationship between the isolated strains and other related bacterial strains found in the GenBank database. The sequences of the isolates were deposited in GenBank and their accession numbers are EU780459, EU780460, EU780461, EU780462, EU780463, and EU780464. |
| Phenol degradation at various NaCl concentrations by the bacterial consortium |
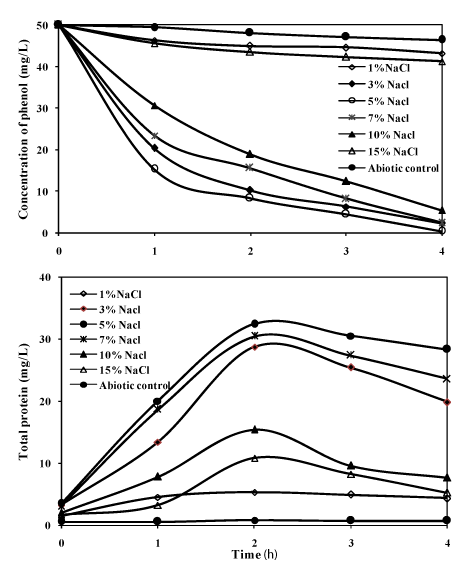
| Initially during the isolation period, the bacterial consortium was studied for its ability to degrade phenol (50mg/L) as sole carbon source and energy at different NaCl concentrations (1%, 3%, 5%, 7%, 10% and 15%). The results on the degradation of phenol at different NaCl concentration are presented in the Figure 4. The growth of the consortium was optimum at 5% NaCl, and when the concentration of sodium chloride was increased to 15%, the growth was inhibited. In earlier reports, with halophilic single strains of bacteria in the degradation of phenol the incubation period for the mineralisation of the substrate was more [7-9,13]. In the present investigation with the bacterial consortium, it was able to completely mineralize the substrate within 96 h at different NaCl concentrations. At 3% NaCl, the degradation efficiency of phenol was 95% and the efficiency reached maximum up to 99% at 5% NaCl, where the protein concentration reached the maximum of 34mg/L. When the NaCl concentration was increased to 7%, 10% and 15%, the degradation decreased to 93%, 89% and 14% respectively. The reduction in the efficiency of degradation of phenol above 10% NaCl may be due to increase in salinity which was supported by a few studies [27-29]. The phenol degradation by the consortium was as low as 10% at 1% NaCl, this was also indicated by the very less protein yield of 6mg/L which emphasizes that the bacterial consortium needed salinity for utilizing phenol as the substrate; this showed that the bacterial strains in the consortium were moderately halophilic in nature. |
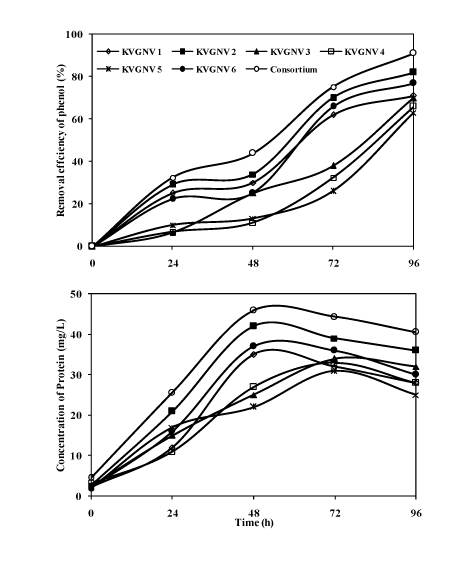
| Comparison of degradation of phenol (300 mg/L) by the bacterial consortium and individual strains |
| The bacterial consortium could grow up to 15% NaCl concentration, however, optimum degradation of phenol was achieved at 5% NaCl where the degradation efficiency was about 99%. This was supported by the few studies [9,30]. Studies of halophilic phenol degradation have been conducted at initial phenol concentrations of 100 mg/L by Hinteregger and Streichsbier [9] and 120 mg/L by Woolard and Irvine [30]. Degradation studies were compared on the utilization of phenol (300 mg/L) by the bacterial strains and the consortium. The degradation of phenol analysed by gas-chromatography showed the retention time of 3.92 min, Figure 5 shows the degradation of phenol by the consortium at 24 hours time interval. Figure 6 shows the degradation of phenol (300 mg/L) by the individual strains and the bacterial consortium. At optimum salinity of 5% NaCl, the bacterial consortium showed about 90% degradation efficiency with the bacterial consortium. With the individual isolates Arthrobacter sp. showed the highest degradation of 88% with protein content of 42 mg/L, while the bacterial consortium was most efficient in degrading phenol with an efficiency of 90% with a protein yield of 46 mg/L at the end of 2rd day. Pseudomonas aeruginosa, Bacillus cereus, and Bacillus licheniformis gave about 77, 71 and 70% of degradation efficiency with the protein yield in the range of 25-37 mg/L. Further phenol degradation efficiency reduced when Halomonas salina and Bacillus pumilus was used individually to 66 and 63%. Based on the results it was inferred that using a bacterial consortium in the degradation of phenol proved to be more efficient in completely removing phenol in 4 days. Finally comparing the abilities of the bacterial strains and the consortium showed that more than 90% degradation efficiency was achieved in the presence of the consortium. The reason might be due to bacterial strains present in the consortium co-exists and helps each other to tolerate phenol, while as individual strains tolerance to the toxic substrate was limited. In treatment of phenol containing wastewater it would be practically feasible to use bacterial consortium as the inoculum as the individual isolates may be less efficient and may suffer from contamination problems. |
| Conclusion |
| The isolated bacterial consortium had six bacterial isolates which were identified as Bacillus cereus, Arthrobacter sp., Bacillus licheniformis, Halomonas salina, Bacillus subtilis and Pseudomonas aeroginosa by 16S rRNA gene sequence analysis and biochemical tests. The results have demonstrated the ability of moderately halophilic bacterial consortium to rapidly degrade phenol 50 mg/L at different salt concentrations where the optimum degradation was achieved at 5% NaCl under aerobic conditions. At optimum salinity of 5% individual isolates and the consortium were studied for degradation of phenol at 300 mg/L, where the consortium was more efficient in the degradation than the individual isolates. Thus these observations provide hope for the establishment of cost effective methods to remediate phenolic wastewater effluents in high salt environments. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
 |
 |
 |
| Figure 4 | Figure 5 | Figure 6 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 18252
- [From(publication date):
October-2010 - Dec 05, 2025] - Breakdown by view type
- HTML page views : 13204
- PDF downloads : 5048
